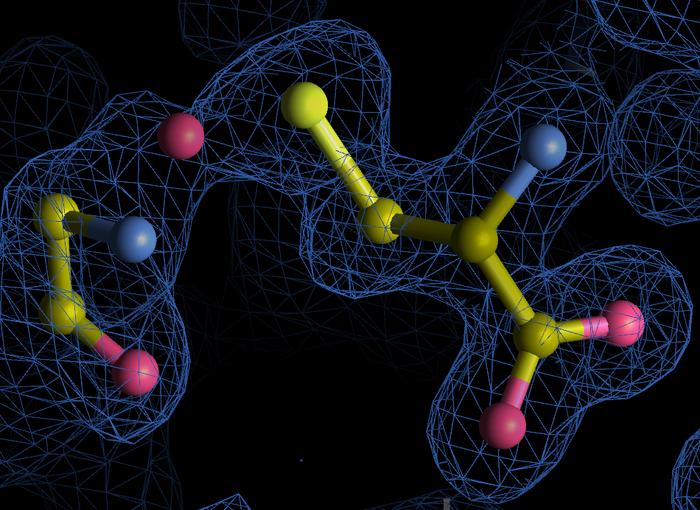
In the sprawling landscape of molecular biology, proteins stand as fundamental pillars supporting virtually every cellular process. Despite the exhaustive research spanning decades, a team of scientists from the University of Göttingen has unveiled previously unknown chemical bonds within protein structures, revealing an extraordinary layer of complexity in protein chemistry. This groundbreaking discovery opens new avenues for understanding how proteins respond to oxidative stress—an often damaging cellular condition marked by the excessive presence of reactive oxygen species (ROS).
Oxidative stress, resulting from an imbalance between reactive oxygen molecules and antioxidant defenses, is known to alter the structure and function of biomolecules. Proteins, being central to cell machinery, are particularly susceptible to oxidative modifications, which can lead to changes in their stability and activity. Until now, the intricate details of how proteins chemically adapt or respond to such stress remained only partially understood. The discovery of novel nitrogen-oxygen-sulphur (NOS) based covalent linkages ushers in a new paradigm for protein biochemistry, highlighting molecular switches invisible to traditional analytical techniques.
The researchers embarked on an ambitious computational re-evaluation of over 86,000 high-resolution protein structures archived within the Protein Data Bank (PDB), the foremost global repository for protein data. Utilizing a cutting-edge algorithm developed in-house, termed SimplifiedBondfinder, the team employed a fusion of machine learning methodologies, quantum mechanical modeling, and rigorous structural refinement algorithms. This innovative pipeline allowed the detection of subtle bond formations—specifically NOS linkages—previously evading conventional structural analyses and experimental validation.
Traditionally, the existence of NOS bonds was recognized between cysteine and serine amino acid residues, with their role briefly characterized within redox biology. However, the Göttingen team’s computational deep dive uncovered NOS linkages in previously uncharted amino acid pairs, including arginine-cysteine and glycine-cysteine combinations. This discovery is particularly notable because it expands the chemical repertoire of post-translational modifications, thereby revealing hitherto unknown molecular mechanisms that proteins can harness under oxidative conditions.
The formation of NOS bonds involves a tri-atomic bridge containing nitrogen, oxygen, and sulfur atoms linking specific amino acid side chains. The chemical implications are profound. These bonds act as reversible molecular switches that confer structural stability and modulate protein function in response to fluctuating oxidative environments. Such mechanisms may fine-tune enzymatic activities, regulate protein-protein interactions, or even protect critical proteins from irreversible oxidative damage.
Dr. Sophia Bazzi, who spearheaded the study at the University of Göttingen’s Institute of Physical Chemistry, emphasized the importance of revisiting established datasets with modern computational tools. “Our findings demonstrate that the Protein Data Bank is not just a static archive but a reservoir teeming with hidden chemistry waiting to be uncovered,” Bazzi remarked. “By coupling machine learning with quantum chemistry, we have charted new chemical territory within proteins that had previously been invisible.”
The implications of this research ripple beyond basic science. Enhanced protein models that incorporate these newly recognized NOS linkages could revolutionize protein engineering efforts. For example, designing enzymes with built-in oxidative stress resilience becomes more feasible when these chemical switches are understood and manipulated. Similarly, drug discovery and synthetic biology stand to benefit from this enriched chemical understanding, potentially accelerating the creation of therapeutics and synthetic biomolecules with superior stability and functionality.
Methodologically, the combination of large-scale computational screening and quantum mechanical validation represents an emerging frontier in structural biology. The SimplifiedBondfinder pipeline was rigorously tested against benchmark data and demonstrated exceptional sensitivity and specificity in detecting otherwise overlooked covalent bonds. This comprehensive re-evaluation not only functions as an analytical breakthrough but also sets a precedent for future explorations into protein structural data, moving the field toward an era of enhanced protein characterization accuracy.
The discovery further underscores the latent value embedded within existing scientific databases. While experimental methodologies continue to advance, computational reinterpretations of archived data can yield transformative insights without the exhaustive need for new laboratory experiments. This synergy between data science and molecular biology maximizes resource utilization, fostering scientific breakthroughs that are both cost-effective and rapid.
Moreover, the chemical novelty of arginine-cysteine and glycine-cysteine NOS bonds hints at diverse biological roles across different protein families. Arginine and glycine are among the most abundant amino acids across proteomes, and their newfound ability to engage in these redox-sensitive linkages broadens the scope of oxidative signaling and regulation. Investigating the functional consequences of these bonds in vivo remains a crucial next step for deciphering their physiological relevance.
In essence, this study heralds a new frontier in our comprehension of protein chemistry, emphasizing that even extensively studied molecules like proteins harbor undiscovered secrets with far-reaching biological consequences. As researchers worldwide begin to incorporate these novel findings into experimental designs and theoretical frameworks, the molecular dance governing life’s vital processes will be better illuminated in its full chemical complexity.
Looking ahead, the development and refinement of similar computational pipelines will be pivotal in uncovering additional atypical bonds and post-translational modifications. This progression promises a more nuanced understanding of proteomic landscapes, potentially unveiling new targets for therapeutic intervention and innovative biomolecular design principles. The work from the University of Göttingen stands as a testament to the profound insights achievable when advanced computational methods meet exhaustive data scrutiny, reshaping the boundaries of molecular biology.
Subject of Research:
Not applicable
Article Title:
Revealing arginine–cysteine and glycine–cysteine NOS linkages by a systematic re-evaluation of protein structures
News Publication Date:
13-May-2025
Web References:
https://doi.org/10.1038/s42004-025-01535-w
References:
Bazzi et al., Communications Chemistry, 2025
Image Credits:
Sophia Bazzi (structural data from the Protein Data Bank, visualization using Coot software)
Keywords:
Protein analysis, Protein interactions, Proteins, Protein activity, Biochemical processes, Enzymology, Life sciences, Cell biology, Chemistry, Physics, Algorithms
Tags: advancements in molecular biologycomputational analysis of protein structureshigh-resolution protein data evaluationmolecular switches in proteinsnitrogen-oxygen-sulphur covalent linkagesnovel protein chemistry discoveriesoxidative stress and protein responseprotein biochemistry breakthroughsprotein structure and stabilityreactive oxygen species effects on proteinsunderstanding protein modificationsUniversity of Göttingen research findings