Credit: BioBlox
BioBlox2D is the result of a collaboration between researchers at Imperial College London and Goldsmiths, University of London. It turns the science of how proteins fit together (or 'dock') with smaller molecules, such as medicines and vitamins, into a Tetris-style puzzle game and quiz. Players manipulate and dock molecules into proteins to score points and earn bonus powers in a race against time.
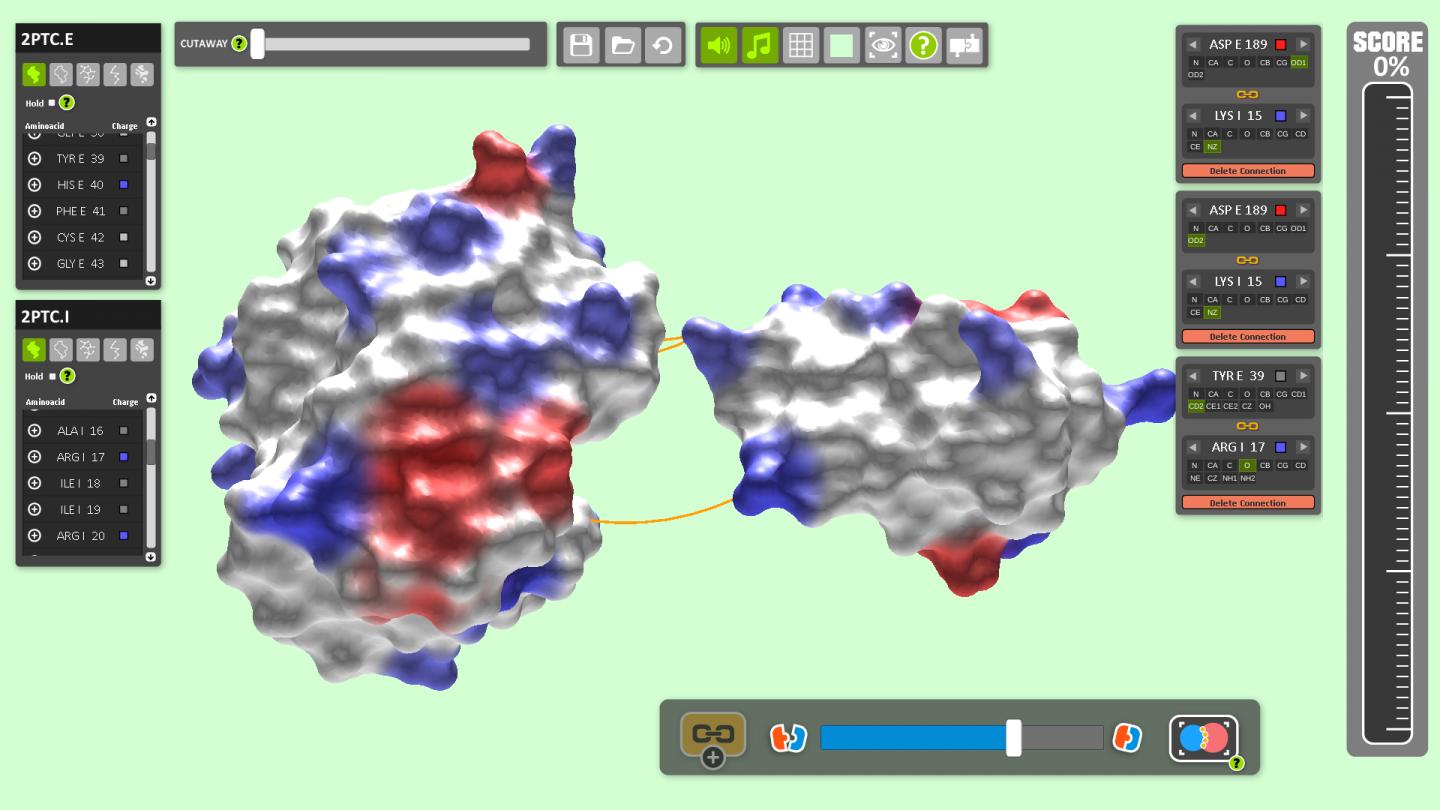
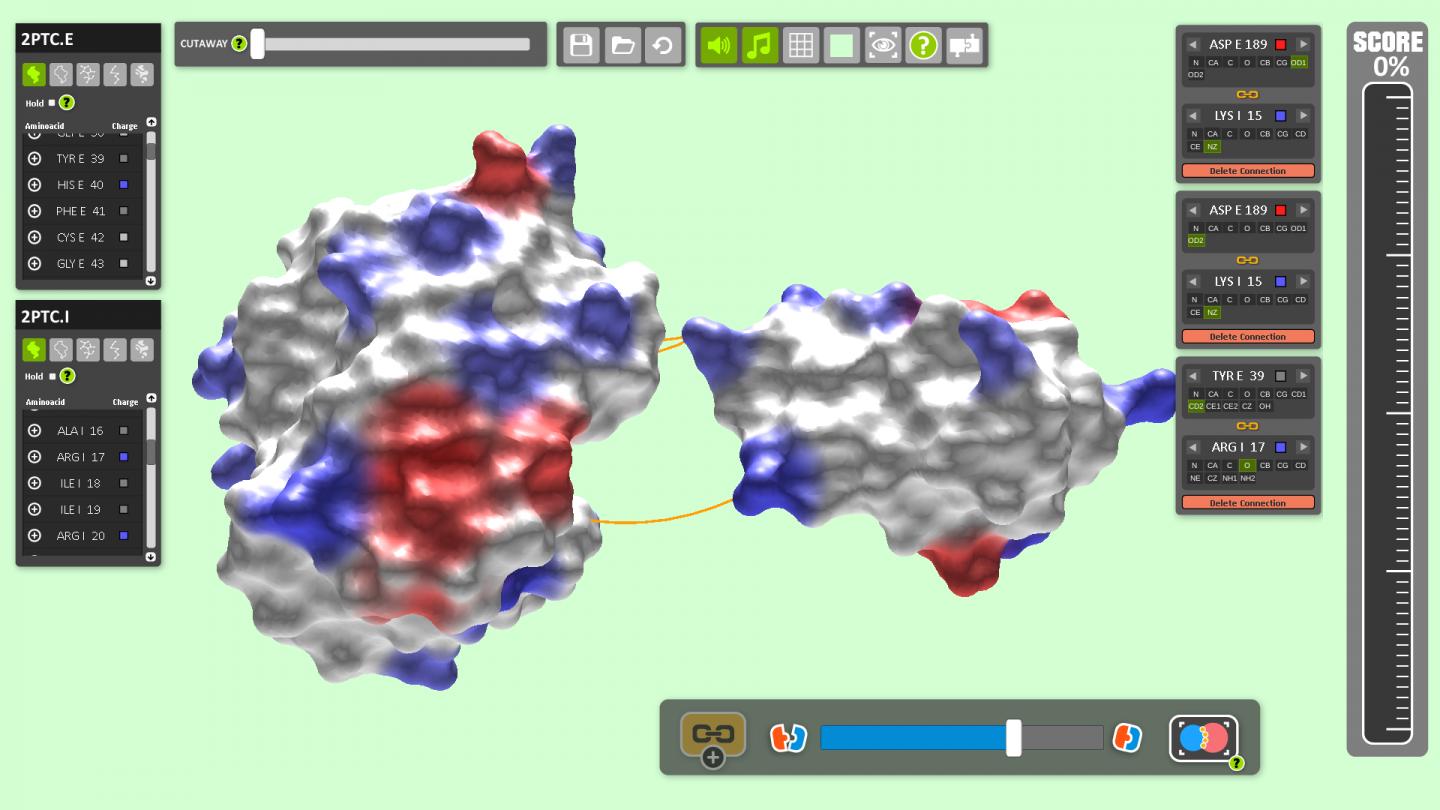
How molecules dock onto proteins is the key to understanding processes in the cell, and in particular to designing new drugs to treat conditions such as cancer and Alzheimer's. The complex 3D forms of such molecules — resembling the bumpy surface of an asteroid full of pits and craters – make understanding how they fit together extremely challenging.
BioBlox2D presents this complex problem in 2D form. The researchers designed the game to be fun but also a valuable educational tool for sharing the importance of protein research. BioBlox2D is free to download from the App store and Google Play.
The researchers aimed to develop a game that is fun to play and helps players to learn about biology. The quiz element of the game asks players to name a biological molecule from its description, for example asking for the molecule that is used by our cells to produce energy — which is later revealed to be glucose. BioBlox2D could therefore be used in schools to teach chemistry and biology.
Professor William Latham, from the Department of Computing at Goldsmiths, and Creative Director of the project, said: "In BioBlox2D we open the world of protein docking to the mass market casual games player, where they have fun playing our puzzle game but at the same time learn about the science."
Professor Michael Sternberg, from the Department of Life Sciences at Imperial and one of project leads, said: "We were inspired by a scientific problem to develop a fun-to-play game where players can experience the challenges of matching both shapes and electrical charges, which is central to how life works."
The researchers say the block-slotting gameplay is given an original twist as players also have to match positively charged blocks with negatively charged ones — a reference to the binding mechanisms of real proteins. Successfully completing rows of blocks unlocks information about the molecules and adds bonuses such as slowing time and automatically completing a level.
The team are also releasing a 3D version at the same time as the 2D version, BioBlox3D, with which they plan to make it possible to crowdsource the protein docking problem through citizen science challenges.
The intention with BioBlox3D is to simulate the protein docking problem with far greater realism in 3D and potentially solve real-world problems. At the moment, the pre-set models in the game come from an existing protein database, but players will soon have the ability to upload their own protein data and experiment in 3D.
Frederic Fol Leymarie, Professor of Computing at Goldsmiths and co-lead on the project, said: "It is hoped this will provide the building blocks for people to create citizen science challenges to, for instance, crowdsource the search for new drug molecules."
###
BioBlox3D is a free and fully accessible web tool, and the demo version is playable on the website.
The project was supported by the Biotechnology and Biological Sciences Research Council (BBSRC).
Media Contact
Hayley Dunning
[email protected]
020-759-42412
@imperialspark
http://www.imperial.ac.uk/press
Original Source
http://www3.imperial.ac.uk/newsandeventspggrp/imperialcollege/newssummary/news_19-6-2017-16-45-33
############
Story Source: Materials provided by Scienmag