The MechMorpho Lab recently published work in PLOS ONE that uses approximate Bayesian computation to help guide experimental approaches
Credit: MechMorpho Lab/University of Pittsburgh
PITTSBURGH (July 9, 2019) … A bioengineering group from the University of Pittsburgh Swanson School of Engineering is bringing the worlds of computational modeling and experimentation closer together by developing a methodology to help analyze the wealth
of imaging data provided by advancements in imaging tools and automated microscopes.
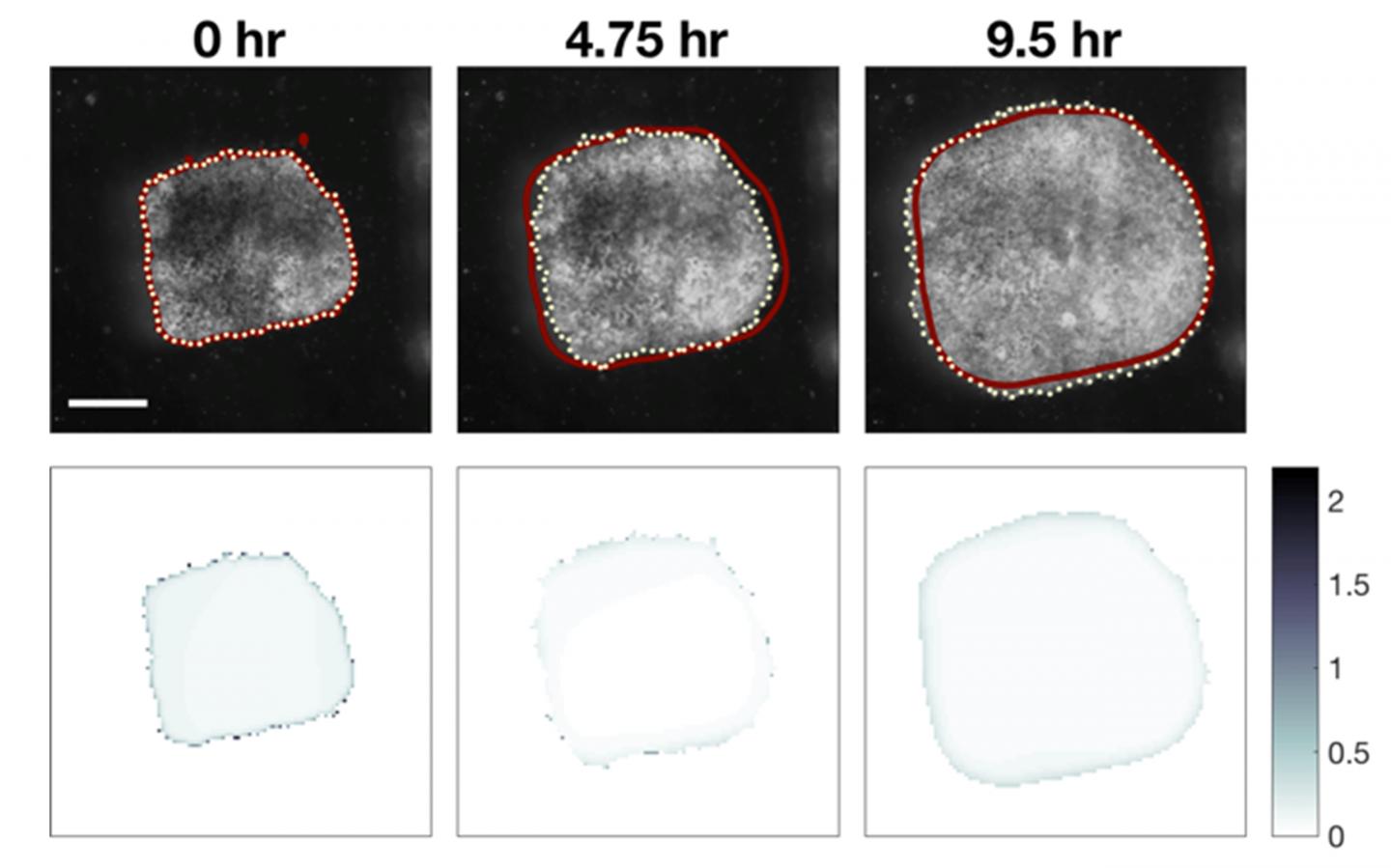
Their study focuses on embryonic tissue spreading, a process that is critical during wound healing and the progression of many diseases. The article, recently published in PLOS ONE (DOI: 10.1371/journal.pone.0218021),
shows how using approximate Bayesian computation (ABC) – a statistical inference method – can help derive useful quantitative information for experimental design.
The work was overseen by Lance Davidson, professor of bioengineering, who runs the MechMorpho Lab in the Swanson School of Engineering. The study was led
by Tracy Stepien, a Pitt mathematics graduate alumnus, and Holley Lynch, a former postdoctoral associate in the MechMorpho Lab.
Davidson’s group cultured tissue from the Xenopus embryo to uncover the mechanical properties behind embryonic morphogenesis – the biological process of an organism developing its shape. During the study, they discovered that small explants spread slower
than larger ones so they began creating modeling approaches to find out why.
They collected time-lapse image sequences over the course a few weeks, but the challenge when integrating modeling with experiments is determining the best set of parameters.
“As models get more complex and the experimental systems produce more data, it is difficult to determine if the chosen parameters are the optimal set,” said Stepien, a postdoctoral associate at the University of Arizona. “This is where Bayesian computation
is useful – for each dataset, you can run the model thousands of times to identify sets of parameters that best match the experiment itself.”
Once the group applied a Bayesian approach to their model, they found that there was no one unique parameter set. Instead, they identified distributions of ‘almost-best’ parameters and then used statistical methods to compare the different distributions.
From the statistical analysis, they predict tissue properties such as force production and adhesion are more likely to vary with initial tissue size than factors such as rates of cell division or shape change.
“Our work provides predictive methods that can help guide more general studies of morphogenesis to better understand how tissue spreading is regulated during development and potentially control spreading during wound healing and cancer,” said Lynch, who
is currently an assistant professor of physics at Stetson University.
According to the research group, these parameters may also prove to be useful for improving models of other experiments.
“Though this paper focused on one specific type of tissue movement, it is more broadly impactful in that it is trying to identify a new way forward for the scientific community,” said Davidson. “Over several years our model went from something simple
to something more rigorous and robust. A paper that originally focused on tissue spreading eventually evolved to demonstrate a framework that very closely integrates modeling and experiment.”
In the short-term, Davidson’s lab will follow-up on the predictions made in this paper and further investigate the biophysics of tissue spreading. In the long-term, the group would like to apply an equally robust statistical approach to other computational
models being developed in the lab.
“We really want to integrate our experimental approaches with computational models, as robust statistically as this approach offers,” said Davidson. “Using the power of computation in conjunction with advanced biomechanical experiments could really impact
our knowledge of disease development and treatment.”
Media Contact
Leah Russell
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




