Credit: Thiel, Kuepfer
A team of researchers has used a computational modeling approach to analyze and compare the toxic effects of fifteen different drugs on the liver, according to a study in PLOS Computational Biology.
Drugs prescribed for various medical conditions can cause harmful liver side effects. Lab experiments with liver cells can help reveal the underlying molecular mechanisms by which these drugs cause liver damage, which could inform better prevention and treatment efforts. However, lab experiments alone cannot reliably predict actual effects in living patients.
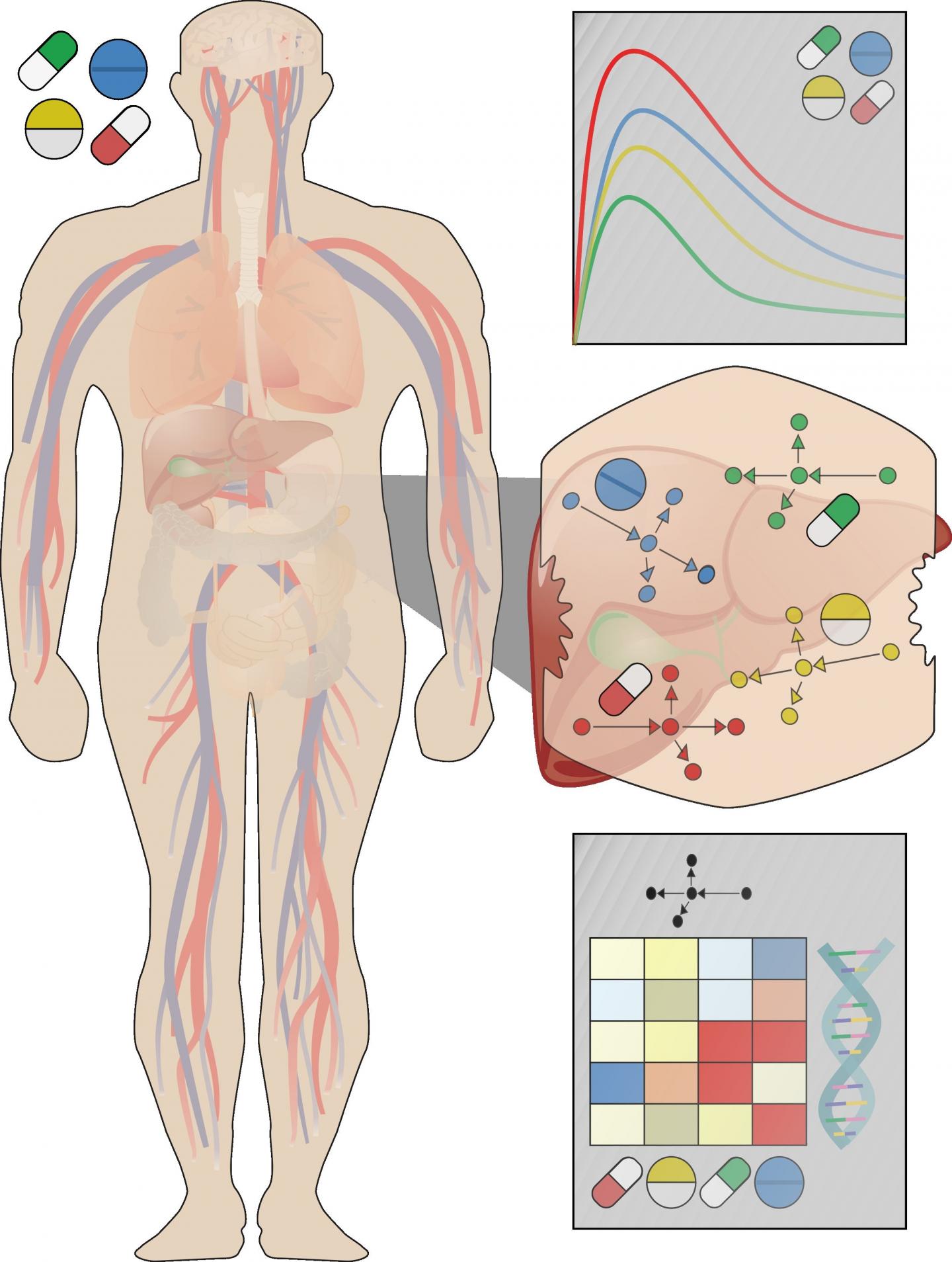
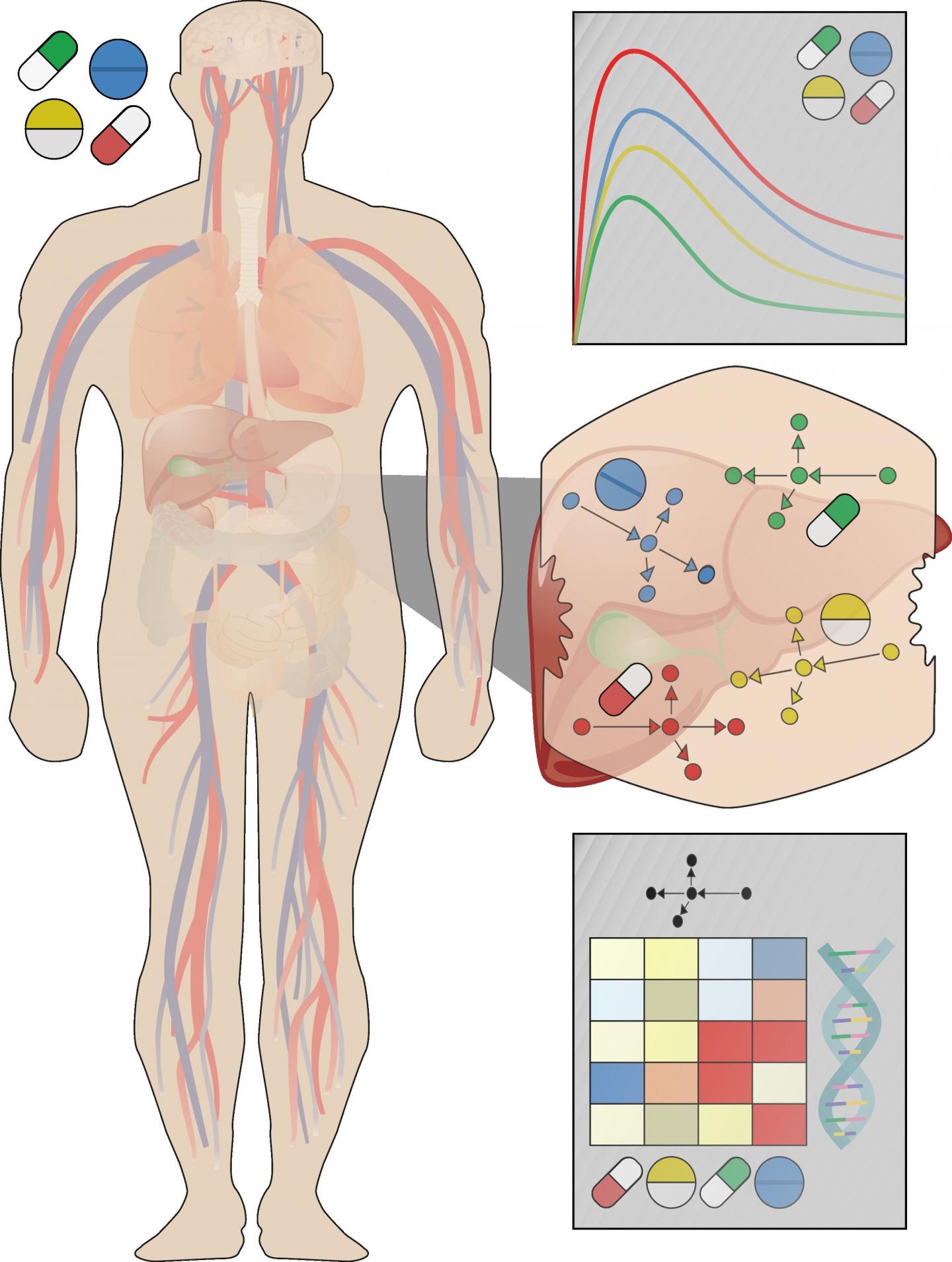
To improve translation of lab data to patients, Christoph Thiel of RWTH Aachen University, Germany, and colleagues recently developed a new strategy that uses computational modeling to simulate how liver cells in the body respond to different doses of different drugs. The approach integrates experimental observations with knowledge of how drugs are distributed and metabolized after they enter the body.
The researchers had previously demonstrated their approach in a proof-of-concept study. In the new study, the approach was applied to simulate and compare the potentially toxic liver effects of fifteen different drugs at clinically relevant doses.
The scientists developed whole-body models to simulate the fate of each drug after ingestion and validated the models using experimental data from scientific literature. These models were then coupled with lab data to predict each drug's effects on the liver at patient level. The researchers found that the drugs fell into different groups that caused similar responses, including which genes would be transcribed in response to toxic doses.
While further validation is required, the method has the potential to lead to faster diagnosis of toxic liver side effects in patients. It could help reveal which gene transcripts could serve as early signs of toxicity and which drug combinations might be particularly dangerous, for both new and existing drugs.
"Consistently applied to the design of clinical development programs, the approach presented has the potential to early identify medical and economic risks of new drugs," says study co-author Lars Kuepfer.
###
In your coverage please use this URL to provide access to the freely available article in PLOS Computational Biology: http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005280
Citation: Thiel C, Cordes H, Fabbri L, Aschmann HE, Baier V, Smit I, et al. (2017) A Comparative Analysis of Drug-Induced Hepatotoxicity in Clinically Relevant Situations. PLoS Comput Biol 13 (2): e1005280. doi:10.1371/journal.pcbi.1005280
Funding: The authors acknowledge financial support by the European Union Seventh Framework Programme HeCaToS (FP7/2007-2013) under the grant agreement no. 602156. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests: LK is an employee of Bayer Technology Services GmbH, the company developing the PBPK modeling tools PK-Sim and MoBi.
Media Contact
Lars Kuepfer
[email protected]
############
Story Source: Materials provided by Scienmag