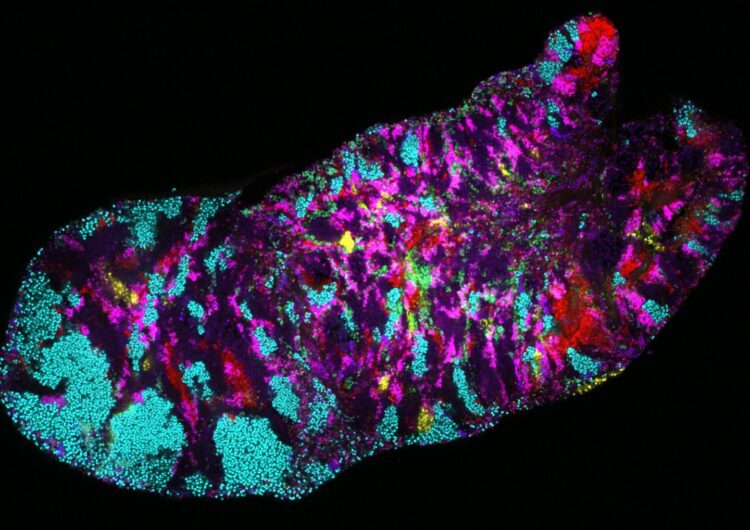
Researchers take a closer look at the genomes of microbial communities in the human mouth
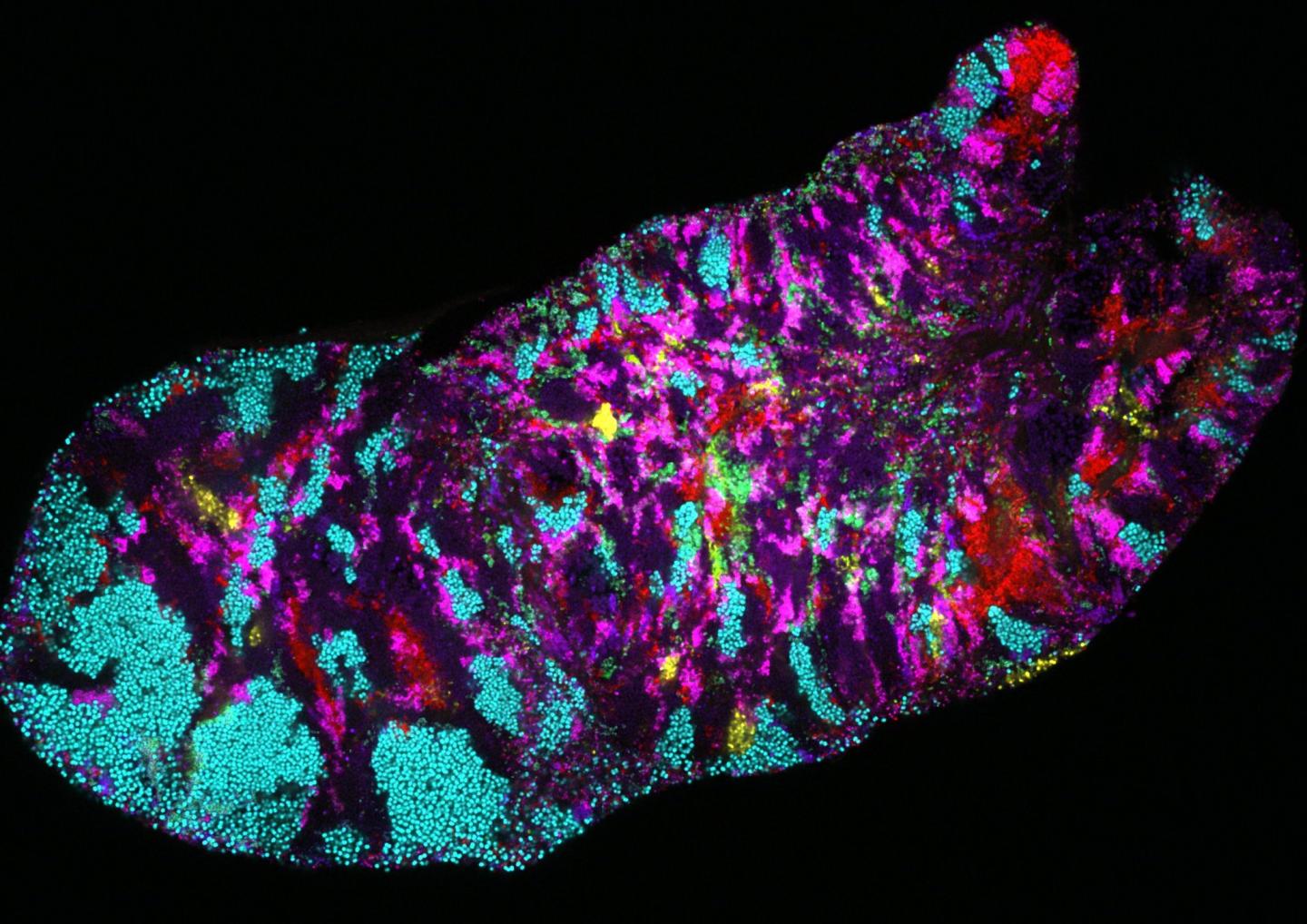
Credit: Photo credit: Jessica Mark Welch, Marine Biological Laboratory.
Bacteria often show very strong biogeography – some bacteria are abundant in specific locations while absent from others – leading to major questions when applying microbiology to therapeutics or probiotics: how did the bacteria get into the wrong place? How do we add the right bacteria into the right place when the biogeography has gotten ‘out of whack’?
These questions, though, have one big obstacle, bacteria are so tiny and numerous with very diverse and complicated populations which creates major challenges to understanding which subgroups of bacteria live where and what genes or metabolic abilities allow them to thrive in these ‘wrong’ places.
In a new study published in Genome Biology researchers led by Harvard University examined the human oral microbiome and discovered impressive variability in bacterial subpopulations living in certain areas of the mouth.
“As microbial ecologists, we are fascinated by how bacteria can seemingly divide up any habitat into various niches, but as humans ourselves, we also have this innate curiosity about how microbes pattern themselves within our bodies,” said lead author Daniel R. Utter, PhD candidate in the Department of Organismic and Evolutionary Biology, Harvard University.
Recent developments in sequencing and bioinformatic approaches have offered new ways to untangle the complexity of bacterial communities. Utter and Colleen Cavanaugh, Edward C. Jeffrey Professor of Biology in the Department of Organismic and Evolutionary Biology, Harvard University, teamed up with researchers at the Marine Biological Laboratory, Woods Hole, University of Chicago, and The Forsyth Institute to apply these state-of-the-art sequencing and analysis approaches to get a better picture of the oral microbiome.
“The mouth is the perfect place to study microbial communities,” according to co-author A. Murat Eren, assistant professor in the Department of Medicine at the University of Chicago. “Not only is it the beginning of the GI tract, but it’s also a very special and small environment that’s microbially diverse enough that we can really start to answer interesting questions about microbiomes and their evolution.”
The mouth contains a surprising amount of site-specific microbes in different areas. For instance, the microbes found on the tongue are very different from the microbes found on the plaque on teeth. “Your tongue microbes are more similar to those living on someone else’s tongue than they are to those living in your throat or on your gums!” said Eren.
The team scoured public databases and downloaded 100 genomes that represented four species of bacteria commonly found in the mouth, Haemophilus parainfluenzae and the three oral species of the genus Rothia, and used them as references to investigate their relatives sampled in hundreds of volunteers’ mouths from the Human Microbiome Project (HMP).
“We used these genomes as a starting point, but quickly moved beyond them to probe the total genetic variation among the trillions of bacterial cells living in our mouths,” said Utter. “Because, at the end of the day, that’s what we’re curious about, not the arbitrary few that have been sequenced.”
Using this recently-developed approach called metapangenomics, which combines pangenomes (the sum of all genes found in a set of related bacteria) with metagenomics (the study of the total DNA coming from all bacteria in a community), allowed the researchers to conduct an in-depth examination of the genomes of the microbes which led to a shocking discovery.
“We found a tremendous amount of variability,” said Utter. “But we were shocked by the patterning of that variability across the different parts of the mouth; specifically, between the tongue, cheek, and tooth surfaces.”
For example, within a single microbe species the researchers found distinct genetic forms that were strongly associated to a single, different site within the mouth. In many cases, the team was able to identify a handful of genes that might explain a particular bacterial group’s specific habitat. Applying metapangenomics the researchers were also able to identify specific ways free-living bacteria in people’s mouths differed from their lab-grown relatives.
“The resolution afforded by these techniques — via the direct comparison of genomes of “domesticated” and “wild” bacteria — allows us to dissect these differences gene by gene,” notes Cavanaugh. “We were also able to identify novel bacterial strains related to, but different than, those we have in culture.”
“Having identified some really strong bacterial candidates that could determine adaptation to a particular habitat, we would like to experimentally test these hypotheses,” said Cavanaugh. These findings could potentially be the key to unlocking targeted probiotics, where scientists could use what’s been learned about each microbe’s habitat’s requirements to engineer beneficial microbes to land in a specified habitat.
“The mouth is so easily accessible that people have been working on bacteria from the mouth for a long time,” said co-author Jessica Mark Welch, associate scientist at the Marine Biological Laboratory.
“Every environment we look at has these really complicated, complex communities of bacteria, but why is that?” said Mark Welch. “Understanding why these communities are so complex and how the different bacteria interact will help us better understand how to fix a bacterial community that’s damaging our health, telling us which microbes need to be removed or added back in.”
This study and others like it can provide new insights on the role of oral microbes in human health. “The ability to identify specific genes behind habitat adaptation has been somewhat of a ‘holy grail’ in microbial ecology,” said Utter. “We are very excited for our contributions in this area!”
###
Article and author details
Daniel R. Utter, Gary G. Borisy, A. Murat Eren, Colleen M. Cavanaugh, Jessica L. Mark Welch. 2020. Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. Genome Biology. DOI: 10.1186/s13059-020-02200-2
Corresponding authors
Daniel R. Utter, [email protected]
Colleen M. Cavanaugh, [email protected]
Support
Support was provided by Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102 and financial contributions from Harvard University and its affiliated academic health care centers). Additional support was provided from NIH Grant DE022586 to G.G.B. Additional support was provided by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE1745303 to D.R.U
Media Contact
Wendy Heywood
[email protected]