Credit: Girirajan Laboratory, Penn State
Genes located in a large chromosomal aberration associated with autism interact with each other to modulate the variable symptoms of the disease, according to new research. A collaborative team led by Penn State researchers tested the role of these genes individually and in tandem by reducing the amount of the genes expressed in a fruit fly model. The research, which appears June 29, 2018 in the journal Nature Communications, demonstrates the utility of the fly as an experimental model for understanding the complex causation of human neurodevelopmental disorders and identifies potential targets for therapeutic treatment.
"As opposed to diseases that are caused by mutations in single genes, autism and other neurodevelopmental disorders have a complex etiology that can be the result of many interacting genes," said Santhosh Girirajan, associate professor of biochemistry and molecular biology and of anthropology at Penn State and the lead author of the paper. "The tools available in flies for genetic manipulation allowed us to quickly test many individual and pairs of genes, so that we can begin to piece together the complexity of their interactions and how that can impact disease symptoms."
Many neurodevelopmental disorders–including autism, schizophrenia, and intellectual disability–are associated with deletions or duplications of relatively large regions of chromosomes that contain many genes. These deletions and duplications result in individuals having a different number of copies of the genes contained with them. This 'copy number variation' can result in developmental defects and neurodevelopmental disorders because an individual may have too much or too little of a particular gene.
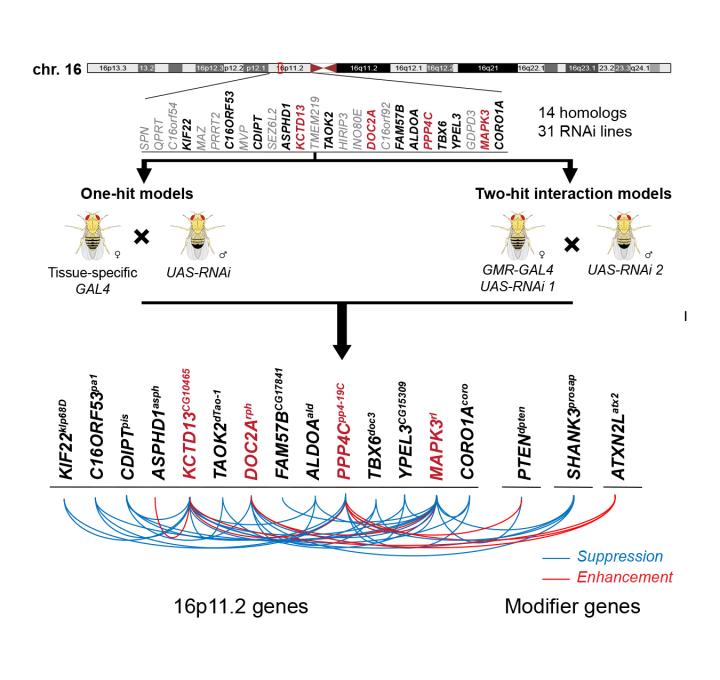
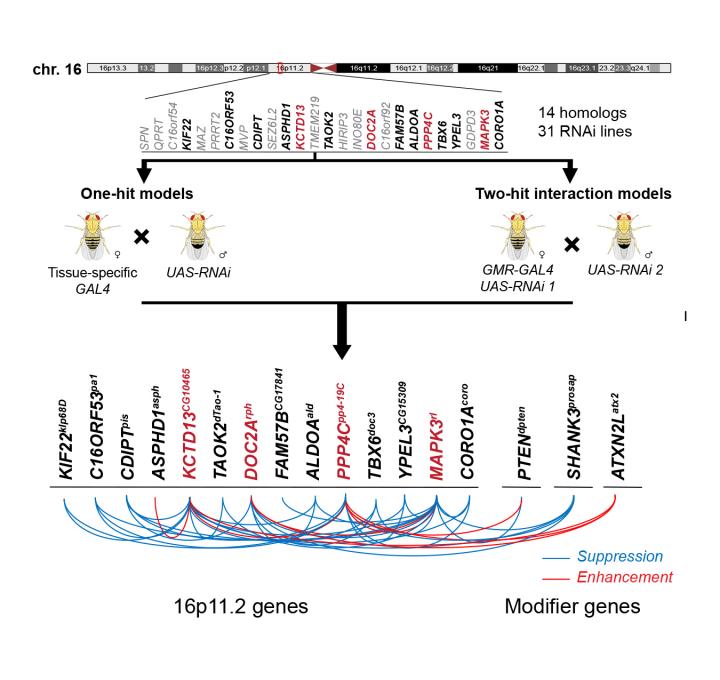
The researchers focused on a large deletion on chromosome 16 that spans over 500,000 base pairs–pairs of As, Ts, Cs, and Gs that make up double-stranded DNA–and was first discovered in individuals with autism. This deletion encompasses 25 individual genes and has subsequently been associated with epilepsy, intellectual disability, and obesity.
"We can sometimes identify a single gene within a large deletion that causes the disease associated with the deletion, but this is rare and usually associated with a specific set of symptoms," said Girirajan. "The deletion on chromosome 16 is associated with many different symptoms that are present to varying degrees–or sometimes not at all–depending on the individual. This led us to believe that there is not a single causative gene, but many that interact in complex ways."
To try to identify how the deletion on chromosome 16 was causing these symptoms, the researchers turned to the fruit fly, Drosophila melanogaster. Flies have been classically used to discover developmental and cellular mechanisms relevant to human biology, and for studying neurodegenerative diseases, sleep disorders and epilepsy. There are also many established tools for genetic manipulation available in flies that allow the researchers to perform experiments much more quickly than in other model organisms, like the mouse.
"Although we only tested the 14 fly counterparts of the 25 human genes, our results suggest a model that can be applied to the overall mechanism of disease caused due to this deletion," said Janani Iyer, a postdoctoral researcher in the Girirajan lab and an author of the paper.
Using a technique called RNA interference (RNAi) in which a short piece of RNA can be designed to reduce the expression of a particular gene in either the entire fly or in a particular tissue, the researchers first tested each of the individual genes in the chromosome 16 deletion that have counterparts in the fly.
"When we tested reducing the expression of individual genes in the fly larva, seven were lethal and we also found other neurodevelopmental effects," said Girirajan. "When we limited the reduction of gene expression to the developing fly eye, we noticed defects connected to cell proliferation."
The researchers then chose the four genes with the most severe effects in the individual tests and tested them in combination with each of the other 13 fly counterparts of the human genes in the chromosome 16 deletion. They also tested the interaction of these four genes with 50 other genes known to play a role in neurodevelopmental disorders from elsewhere in the genome. In the pairwise experiments, the researchers found that the symptoms that appeared in individual tests were sometimes made more severe, sometimes reduced, and sometimes unaffected. This suggested that multiple genes within the deletion likely interact with each other, potentially contributing to the variability in symptoms seen in individuals with the deletion.
"Based on our results, we suggest a new model for the pathogenicity of the chromosome 16 deletion, and other large regions of copy number variation in the genome," said Girirajan. "We must consider the interactions among many genes to understand the variable symptoms associated with these large deletions, rather than looking for single causative genes. Understanding this will help us identify appropriate targets for possible treatment of these neurodevelopmental disorders."
###
In addition to Girirajan, the research team includes Janani Iyer, Mayanglambam Dhruba Singh, Matthew Jensen, Payal Patel, Lucilla Pizzo, Emily Huber, Alexis T. Weiner, Komal Vadodaria, Alexis Kubina, Qingyu Wang, Abigail Talbert, Sneha Yennawar, and Melissa M. Rolls at Penn State; Haley Koerselman and J. Robert Manak at the University of Iowa; Paola Lepanto and Jose Badano at Institut Pasteur de Montevideo in Uruguay; and Arjun Krishnan at the Michigan State University.
The research was supported by a Basil O'Connor Award from the March of Dimes Foundation, the U.S. National Institutes of Health, a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation, and the Penn State Huck Institutes of the Life Sciences.
Media Contact
Sam Sholtis
[email protected]
814-865-1390
@penn_state
http://live.psu.edu
Original Source
http://science.psu.edu/news-and-events/2018-news/Girirajan6-2018 http://dx.doi.org/10.1038/s41467-018-04882-6