Credit: Caltech
The discovery of antibiotics in the early part of the 20th century changed modern medicine. Simple infections that previously killed people became easy to treat. Antibiotics' ability to stave off infections made possible routine surgeries, organ transplants, and chemotherapy for the treatment of cancer.
But because of overuse and misuse, antibiotics are losing their effectiveness. Many species of bacteria have evolved resistance to commonly used antibiotics and multidrug-resistant bacteria–so-called superbugs–have emerged, plaguing hospitals and nursing homes. Last month, the World Health Organization issued a dire warning: The world is running out of antibiotics.
A new test developed at Caltech that identifies antibiotic-resistant bacteria in as little as 30 minutes could help turn the tide by allowing medical professionals to better choose which antibiotics to treat an infection with. A paper describing the method appears in the October 4 issue of Science Translational Medicine.
When doctors treat patients with bacterial infections, they often skip over first-line antibiotics like methicillin or amoxicillin–drugs that bacteria are more likely to be resistant to–and go straight for stronger second-line antibiotics, like ciprofloxacin. This practice increases the chance that the treatment will be effective, but it is not ideal. That's because the increased use of second-line antibiotics makes it more likely that bacteria also will become resistant to these stronger drugs.
"Right now, we're overprescribing, so we're seeing resistance much sooner than we have to for a lot of the antibiotics that we would otherwise want to preserve for more serious situations," says Nathan Schoepp, a Caltech graduate student and co-author of the study.
The problem is that there has not been a quick and easy way for a doctor to know if their patient's infection is resistant to particular antibiotics. To find out, the doctor would have to send a sample to a testing lab, and wait two to three days for an answer.
"Therapies are driven by guidelines developed by organizations like the World Health Organization or the Centers for Disease Control and Prevention without knowing what the patient actually has, because the tests are so slow," says Rustem Ismagilov, Caltech's Ethel Wilson Bowles and Robert Bowles Professor of Chemistry and Chemical Engineering and director of the Jacobs Institute for Molecular Engineering for Medicine. "We can change the world with a rapid test like this. We can change the way antibiotics are prescribed."
Ismagilov, Schoepp, Caltech graduate student Travis Schlappi, who is also a co-author, and their fellow researchers aimed to develop a test that could be completed during a single visit to the doctor's office. They focused on one of the most common types of infections in humans, urinary tract infections (UTIs), which 50 percent of women contract during their lifetimes. UTIs result in eight million doctor visits and one million ER visits each year in the United States alone.
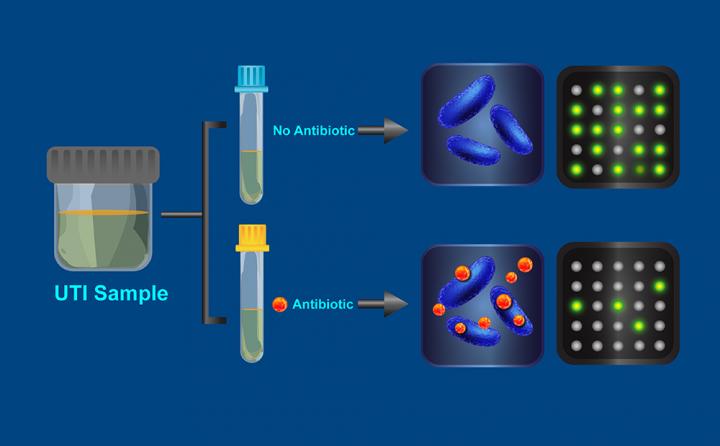
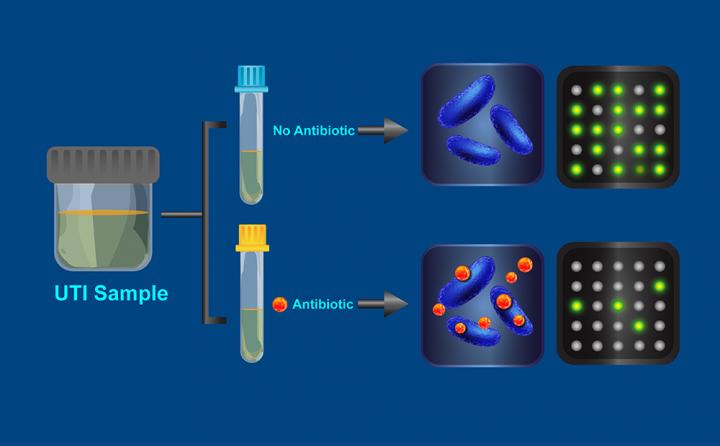
The researchers' new test works like this: A sample of urine (which may contain bacteria) collected from a patient with a UTI is divided into two parts. One part is exposed to an antibiotic for 15 minutes, while the other part incubates without antibiotics. The bacteria from each sample then are broken open (lysed) to release their cellular contents, which are run through a process that combines a detection chemistry technique called digital real-time loop-mediated isothermal amplification, or dLAMP, with a device called a SlipChip (SlipChips are a previous invention of Ismagilov and his Caltech colleagues). This combination replicates specific DNA markers so they can be imaged and individually counted as discrete fluorescent spots appearing on the chip.
The test operates on the principle that typical bacteria will replicate their DNA (in preparation for cellular division) less well in an antibiotic solution, resulting in the presence of fewer DNA markers. However, if the bacteria are resistant to the antibiotic, their DNA replication will not be hampered and the test will reveal similar numbers of DNA markers in both the treated and untreated solutions.
When used on 54 samples of urine from patients with UTIs caused by the bacteria Escherischia coli, the test results had a 95 percent match with those obtained using the standard two-day test, which is considered the gold standard for accuracy.
Ismagilov and Schoepp plan to begin running the test on other types of infectious bacteria to see how well it performs. They also hope to tweak the testing procedures to work with blood samples. Blood infections are more difficult to test because the bacteria are present in much lower numbers than they are in urine, but such a test could help reduce mortality from blood-borne infections, which can turn fatal if not treated quickly.
###
The paper is titled "Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples." Other co-authors include graduate students Matthew S. Curtis of Caltech; Slava S. Buktovich (BS '17); and Shelley Miller and Romney M. Humphries of UCLA. The research was supported by the Defense Advanced Research Projects Agency, the National Institutes of Health, the Burroughs Wellcome Fund, and the Jacobs Institute for Molecular Engineering for Medicine. SlipChip Corp., of which Ismagilov is a founder and shareholder, provided some of the chips used in the study.
Media Contact
Emily Velasco
[email protected]
626-536-6915
@caltech
http://www.caltech.edu
Original Source
https://www.caltech.edu/news/caltech-researchers-create-test-reveals-antibiotic-resistant-bacteria-30-minutes-79943 http://dx.doi.org/10.1126/scitranslmed.aal3693