Credit: Paul Tar
A machine learning approach for assessing images of the craters and dunes of Mars, which was developed at The University of Manchester, has now been adapted to help scientists measure the effects of treatments on tumours.
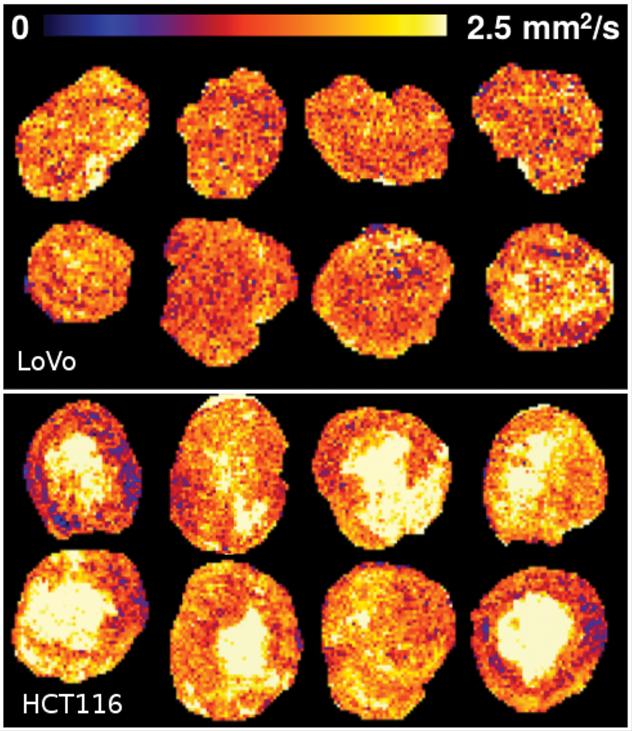
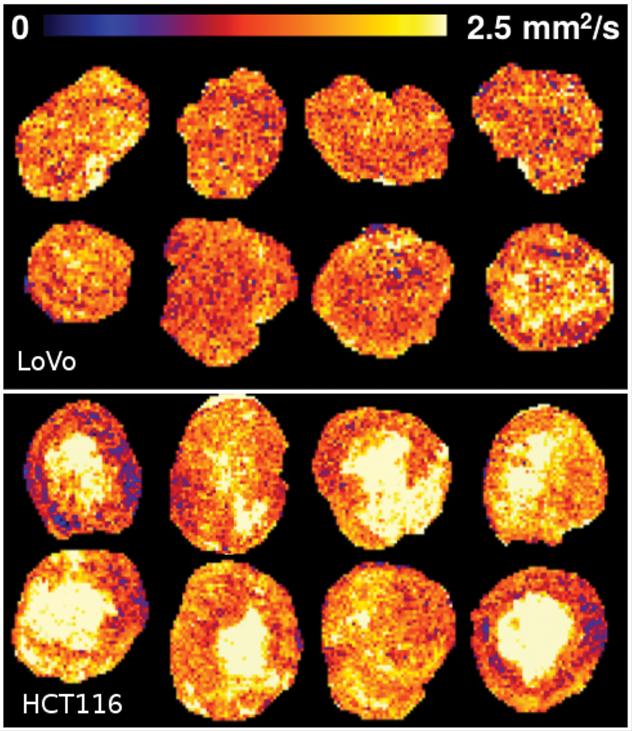
Because tumours are not uniform and different parts of them change at varying speeds, it is difficult for researchers to see what effects their treatments are having against a background of changes that would happen anyway.
Typically, to obtain meaningful results scientists have to look at average changes in tumours using many samples, often in animals. With conventional statistical methods, it can be difficult to assess the effects of treatment on individuals, as would be required for personalised medicine.
The machine learning technique was developed at Manchester to help planetary scientists map features on planets such as Mars. It was designed to better understand the errors and uncertainties of observations, thereby enabling researchers to present their findings with confidence.
The Manchester team, from the Division of Informatics, Imaging & Data Sciences worked in collaboration with Dr James O'Connor, Head of Imaging within the Manchester Cancer Research Centre on studies of lab mice. They applied their machine learning technique, called Linear Poisson Modelling, to the samples and were able to demonstrate a four-fold increase in the precision of tumour change measurements that detected the beneficial effects of cancer therapies.
Dr Neil Thacker, from the University's Division of Informatics, Imaging & Data Sciences, said: "The results of this study show that we can present findings which researchers can be much more certain of. This means you can get the same quality of data from one sample instead of 16."
This has important implications for research, meaning that instead of using 16 mice, in some studies only one is needed. This could help reduce the use of lab mice in medical research. It also opens up the potential for this technique to be used in patients by quickly and confidently identifying if drugs are having a specific effect on their tumours."
Linear Poisson Modelling works by learning patterns within data and how they can change. Unlike other machine learning methods, such as the popular Deep Learning, it can also assess the effects of errors in data, providing as an additional output predictions of how precise its results are. The improved modelling of data also means that fewer samples are needed to provide highly accurate results.
Dr Paul Tar, who co-developed the method during his PhD project, added: "This technique is all about making the most of 'small data', which is common in medical studies where it is difficult to obtain large numbers of samples. Researchers use charitable or public money, so it is important that they use it in the most efficient way possible, something which this technique allows."
Dr James O'Connor, a Cancer Research UK advanced clinician scientist, said: "Every person's cancer is unique, which can make treating the disease challenging as a drug that works for one patient might not work for someone else. That's why we're increasingly looking at finding new ways to make treatment more personal, and this innovative work could be a step towards that goal. The next step will be further research to find out if that's the case, and to help uncover this method's potential."
###
The paper, 'A new method for the high-precision assessment of tumor changes in response to treatment,' has been published in the journal BioInformatics. https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/bty115/4934935
The paper 'Automated quantitative measurements and associated error covariances for planetary image analysis', (advances in Space Research, 2015) is available here: https://www.sciencedirect.com/science/article/pii/S0273117715002549
The project was funded by the Leverhulme Trust, the Royal College of Radiologists and Cancer Research UK. Funding for Dr Tar's PhD was provided by the Science and Technology Facilities Council.
Media Contact
Jamie Brown
[email protected]
44-161-275-8383
@UoMNews
http://www.manchester.ac.uk
Related Journal Article
http://dx.doi.org/10.1093/bioinformatics/bty115