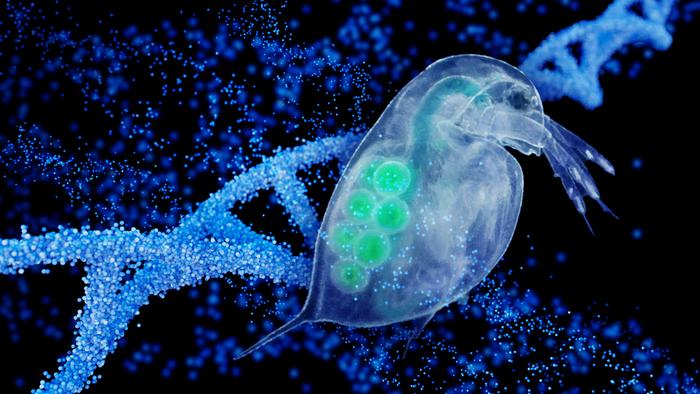
In new research, Arizona State University scientists and their colleagues investigated genetic changes occurring in a naturally isolated population of the water flea, Daphnia pulex. This tiny crustacean, barely visible to the naked eye, plays a crucial role in freshwater ecosystems and offers a unique window into natural selection and evolution.
Credit: Graphic by Jason Drees
In new research, Arizona State University scientists and their colleagues investigated genetic changes occurring in a naturally isolated population of the water flea, Daphnia pulex. This tiny crustacean, barely visible to the naked eye, plays a crucial role in freshwater ecosystems and offers a unique window into natural selection and evolution.
Their findings, reported in the current issue of the journal Proceedings of the National Academy of Sciences (PNAS), rely on a decade of research. Using advanced genomic techniques, the research team analyzed DNA samples from nearly 1,000 Daphnia.
They discovered that the strength of natural selection on individual genes varies significantly from year to year, maintaining variation and potentially enhancing the ability to adapt to future changing environmental conditions by providing raw material for natural selection to act on.
In seemingly stable environments, there is significant fluctuation in the frequency of gene variants known as alleles at specific chromosomal regions over time, even if the overall strength of selection remains near zero on average over many years. This suggests that such genetic variation allows populations to remain adaptable to environmental changes.
“This study has, for the first time, given us a glimpse into the kinds of temporal changes in gene frequencies that occur even in seemingly constant environments, a sort of ongoing churn of genetic variation distributed across the genome,” says Michael Lynch, lead author of the new study.
Lynch is the director of the Biodesign Center for Mechanisms of Evolution and professor in the School of Life Sciences at ASU. Additional researchers on the study include colleagues from ASU, Central China Normal University and the University of Notre Dame.
The power of selection
Daphnia, a form of zooplankton, have fascinated biologists for centuries due to their crucial role in aquatic ecosystems and ability to adapt to environmental stressors. In addition to their value for multigenerational genetic research, Daphnia are widely used model organisms for freshwater toxicity testing because they have a rapid asexual reproductive cycle and are sensitive to various environmental pollutants.
The tiny creatures are a vital food source for fish and help keep algae growth in check. Their ability to adapt quickly to environmental changes could hold clues for how other species — including those important to human food supplies — might respond to pollution, climate change and other human-induced stressors.
Most of the sites examined on the Daphnia genome were shown to experience changing selection pressures over the study period. On average, these pressures tend to balance out to have little overall effect, meaning that no single direction of selection consistently dominates over time. Instead, the genetic advantages or disadvantages of specific traits change from one period to the next.
These findings challenge the traditional belief that measuring genetic diversity (the range of different traits in a population) and genetic divergence (the differences between populations) can easily show how natural selection is consistently operating. Instead, natural selection seems to operate with greater subtlety and complexity than previously thought.
Rethinking genetic variation
The study breaks new ground by pinpointing when and where selection pressures occur within the genome. Other than traits known to be strongly influenced by natural selection, there is little information on how allele frequencies change over time in natural populations.
The multiyear, genome-wide analysis of nearly 1,000 genetic samples from a Daphnia pulex population shows that most genetic sites experience varying selection, with an average effect close to zero, indicating little consistent selection pressure over different times and selection spread across many genomic regions.
These findings challenge the usual understanding of genetic diversity and divergence as indicators of random genetic drift and selection intensity.
Variation and survival
The observed patterns of selection on various gene sites provide a mechanism for maintaining genetic diversity, which is essential for rapid adaptation. The study also revealed that genes located near each other on chromosomes tend to evolve in a coordinated manner. This linkage allows beneficial combinations of gene variants to be inherited together, potentially accelerating the adaptation process.
This effect could help explain how species sometimes adapt faster than scientists would normally expect. On the other hand, the same phenomenon may result in deleterious alleles being swept to higher frequencies by linked beneficial alleles, reducing the overall efficiency of selection in some cases.
The study shows that evolution is more dynamic and complex than previously appreciated. The environment’s influence on genes changes frequently, possibly helping species keep the genetic variety needed to adapt to future conditions. This new understanding may prompt scientists to rethink how they study evolution in the wild.
While the study focused on Daphnia pulex, the findings may have implications for understanding how other species might respond to rapid environmental changes, including those driven by human activities, such as pollution and climate change. Assessing the stability of allele frequencies in more stable environments is an important preliminary step. Such studies are critical, as laboratory experiments alone cannot duplicate the complexity of environmental influences acting on wild populations.
Further, understanding how Daphnia evolve may provide insights into the resilience of entire ecosystems. This knowledge could help researchers predict and potentially mitigate the impacts of environmental changes on biodiversity and food webs.
As the world grapples with an accelerating environmental crisis, studies like this one provide crucial insights into nature’s capacity for resilience and adaptation. By continuing to study these tiny creatures, the scientists hope to better understand the fundamental mechanisms of evolution and apply these lessons to broader ecological and conservation efforts.
Journal
Proceedings of the National Academy of Sciences
DOI
10.1073/pnas.2307107121
Method of Research
Experimental study
Subject of Research
Cells
Article Title
The genome-wide signature of short-term temporal selection
Article Publication Date
3-Jul-2024




