The microscope uses a combination of hardware and software-based adaptive optics to reconstruct object image
Credit: IBS
Non-invasive microscopic techniques such as optical coherence microscopy and two-photon microscopy are commonly used for in vivo imaging of living tissues. When light passes through turbid materials such as biological tissues, two types of light are generated: ballistic photons and multiply scattered photons. The ballistic photons travel straight through the object without experiencing any deflection and hence is used to reconstruct the object image. On the other hand, the multiply scattered photons are generated via random deflections as the light passes through the material and show up as speckle noise in the reconstructed image. As the light propagates through increasing distances, the ratio between multiply scattered and ballistic photons increases drastically, thereby obscuring the image information. In addition to the noise generated by the multiply scattered light, optical aberration of ballistic light also causes contrast reduction and image blur during the image reconstruction process.
Bone tissues in particular have numerous complex internal structures, which cause severe multiple light scattering and complex optical aberration. When it comes to optical imaging of the mouse brain through an intact skull, the fine structures of the nervous system are hard to visualize due to strong speckle noise and image distortion. This is problematic in neuroscience research, where the mouse is widely used as a model organism. Due to the limitation of the currently used imaging techniques, the skull has to be removed or thinned to microscopically investigate the neural networks of brain tissues underneath.
Hence other solutions have been suggested to achieve deeper imaging of living tissues. For example, three-photon microscopy has been successfully used to image neurons beneath the mouse skull in recent years. However, three-photon microscopy is limited by a low laser repetition rate as it employs an excitation window in the infrared range, which can damage the living tissue during in vivo imaging. It also has excessive excitation power, which means photobleaching is more extensive in comparison to the two-photon approach.
Recently, a research team led by Prof. CHOI Wonshik at the Center for Molecular Spectroscopy and Dynamics within the Institute of Basic Science (IBS) in Seoul, South Korea made a major breakthrough in deep-tissue optical imaging. They developed a novel optical microscope that can image through an intact mouse skull and acquire a microscopic map of neural networks in the brain tissues without losing spatial resolution.
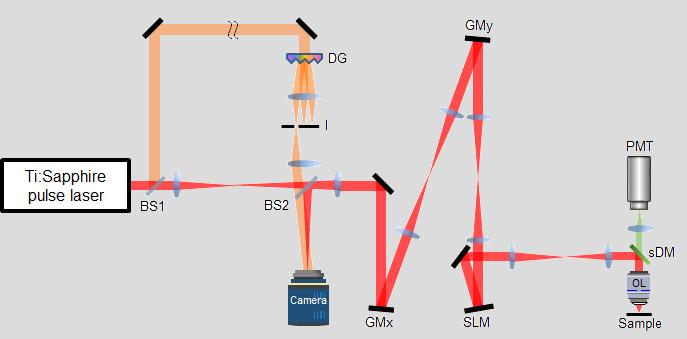
This new microscope is termed as a reflection matrix microscope, and it combines the powers of both hardware and computational adaptive optics (AO), which is a technology originally developed for ground-based astronomy to correct optical aberrations. While conventional confocal microscope measures reflection signal only at the focal point of illumination and discard all out of focus light, the reflection matrix microscope records all the scattered photons at positions other than the focal point. The scattered photons are then computationally corrected using a novel AO algorithm called closed-loop accumulation of single scattering (CLASS), which the team developed back in 2017. The algorithm exploits all scattered light to selectively extract ballistic light and correct severe optical aberration. Compared to the most conventional AO microscopy systems, which require bright point-like reflectors or fluorescent objects as guide stars similarly to the use of AO in astronomy, the reflection matrix microscope works without any fluorescent labeling and without depending on the target’s structures. In addition, the number of aberration modes that can be corrected is more than 10 times greater than that of the conventional AO systems.
The reflection matrix microscope has a great advantage in that it can be directly combined with a conventional two-photon microscope that is already widely used in the life science field. To remove the aberration experienced by the excitation beam of the two-photon microscope, the team deployed hardware-based adaptive optics within the reflection matrix microscope to counteract the aberration of the mouse skull. They showcased the capabilities of the new microscope by taking two-photon fluorescence images of a dendritic spine of a neuron behind the mouse skull, with a spatial resolution close to the diffraction limit. Normally a conventional two-photon microscope cannot resolve the delicate structure of the dendrite spine without removing the brain tissue from the skull entirely. This is a highly significant achievement, as the South Korean group demonstrated the first high-resolution imaging of neural networks through an intact mouse skull. This means that it is now possible to investigate the mouse brain in its most native states.
Research professor YOON Seokchan and graduate student LEE Hojun, who conducted the study, said, “By correcting the wavefront distortion, we can focus light energy on the desired location inside the living tissue.” “Our microscope allows us to investigate fine internal structures deep within living tissues that cannot be resolved by any other means. This will greatly aid us in early disease diagnosis and expedite neuroscience research.”
The researchers set their next research direction to minimize the form factor of the microscope and increase its imaging speed. The goal is the development of a label-free reflective matrix microscope with high imaging depth for use in clinics.
Vice director CHOI Wonshik said, “Reflection matrix microscope is the next-generation technology that goes beyond the limitations of conventional optical microscopes. This will allow us to widen our understanding of the light propagation through scattering media and expand the scope of applications that an optical microscope can explore.”
###
This research was published in the online edition of Nature Communications (IF 11.878) on November 12th.
Media Contact
Wonshik Choi
[email protected]
Related Journal Article
http://dx.




