Rutgers researchers retraced the evolution of enzymes over billions of years
Credit: Vikas Nanda/Rutgers University
Rutgers researchers have discovered the origins of the protein structures responsible for metabolism: simple molecules that powered early life on Earth and serve as chemical signals that NASA could use to search for life on other planets.
Their study, which predicts what the earliest proteins looked like 3.5 billion to 2.5 billion years ago, is published in the journal Proceedings of the National Academy of Sciences.
The scientists retraced, like a many thousand piece puzzle, the evolution of enzymes (proteins) from the present to the deep past. The solution to the puzzle required two missing pieces, and life on Earth could not exist without them. By constructing a network connected by their roles in metabolism, this team discovered the missing pieces.
“We know very little about how life started on our planet. This work allowed us to glimpse deep in time and propose the earliest metabolic proteins,” said co-author Vikas Nanda, a professor of Biochemistry and Molecular Biology at Rutgers Robert Wood Johnson Medical School and a resident faculty member at the Center for Advanced Biotechnology and Medicine. “Our predictions will be tested in the laboratory to better understand the origins of life on Earth and to inform how life may originate elsewhere. We are building models of proteins in the lab and testing whether they can trigger reactions critical for early metabolism.”
A Rutgers-led team of scientists called ENIGMA (Evolution of Nanomachines in Geospheres and Microbial Ancestors) is conducting the research with a NASA grant and via membership in the NASA Astrobiology Program. The ENIGMA project seeks to reveal the role of the simplest proteins that catalyzed the earliest stages of life.
“We think life was built from very small building blocks and emerged like a Lego set to make cells and more complex organisms like us,” said senior author Paul G. Falkowski, ENIGMA principal investigator and a distinguished professor at Rutgers University-New Brunswick who leads the Environmental Biophysics and Molecular Ecology Laboratory. “We think we have found the building blocks of life – the Lego set that led, ultimately, to the evolution of cells, animals and plants.”
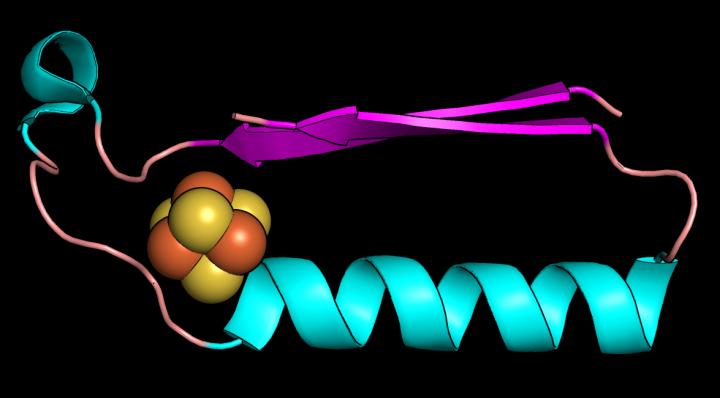
The Rutgers team focused on two protein “folds” that are likely the first structures in early metabolism. They are a ferredoxin fold that binds iron-sulfur compounds, and a “Rossmann” fold, which binds nucleotides (the building blocks of DNA and RNA). These are two pieces of the puzzle that must fit in the evolution of life.
Proteins are chains of amino acids and a chain’s 3D path in space is called a fold. Ferredoxins are metals found in modern proteins and shuttle electrons around cells to promote metabolism. Electrons flow through solids, liquids and gases and power living systems, and the same electrical force must be present in any other planetary system with a chance to support life.
There is evidence the two folds may have shared a common ancestor and, if true, the ancestor may have been the first metabolic enzyme of life.
###
The lead author is Hagai Raanan, a former post-doctoral associate in the Environmental Biophysics and Molecular Ecology Laboratory. Rutgers co-authors include Saroj Poudel, a post-doctoral associate, and Douglas H. Pike, a doctoral student in the ENIGMA project.
Media Contact
Todd Bates
[email protected]
848-932-0550




