Credit: Tao Laboratory/Rice University
The structure of a protein key to the survival and spread of a virus that affects salmon could inform strategies to treat the flu in humans, according to scientists at Rice University.
The Rice lab of structural biologist Yizhi Jane Tao produced the first full-length structure of the matrix protein found in an orthomyxovirus that causes anemia in Atlantic salmon. Because the protein's structure and function are so similar to what has been found so far in influenza viruses, Tao expects they will be useful in determining the mechanisms of human viruses as well.
"This virus and all the influenza viruses belong to the same family, so they assemble in a very similar way," she said. "Although this protein is from a fish-infecting virus, it will give us insight as to how the matrix proteins support the assembly of other viruses."
The discovery is detailed this month in the Proceedings of the National Academy of Sciences.
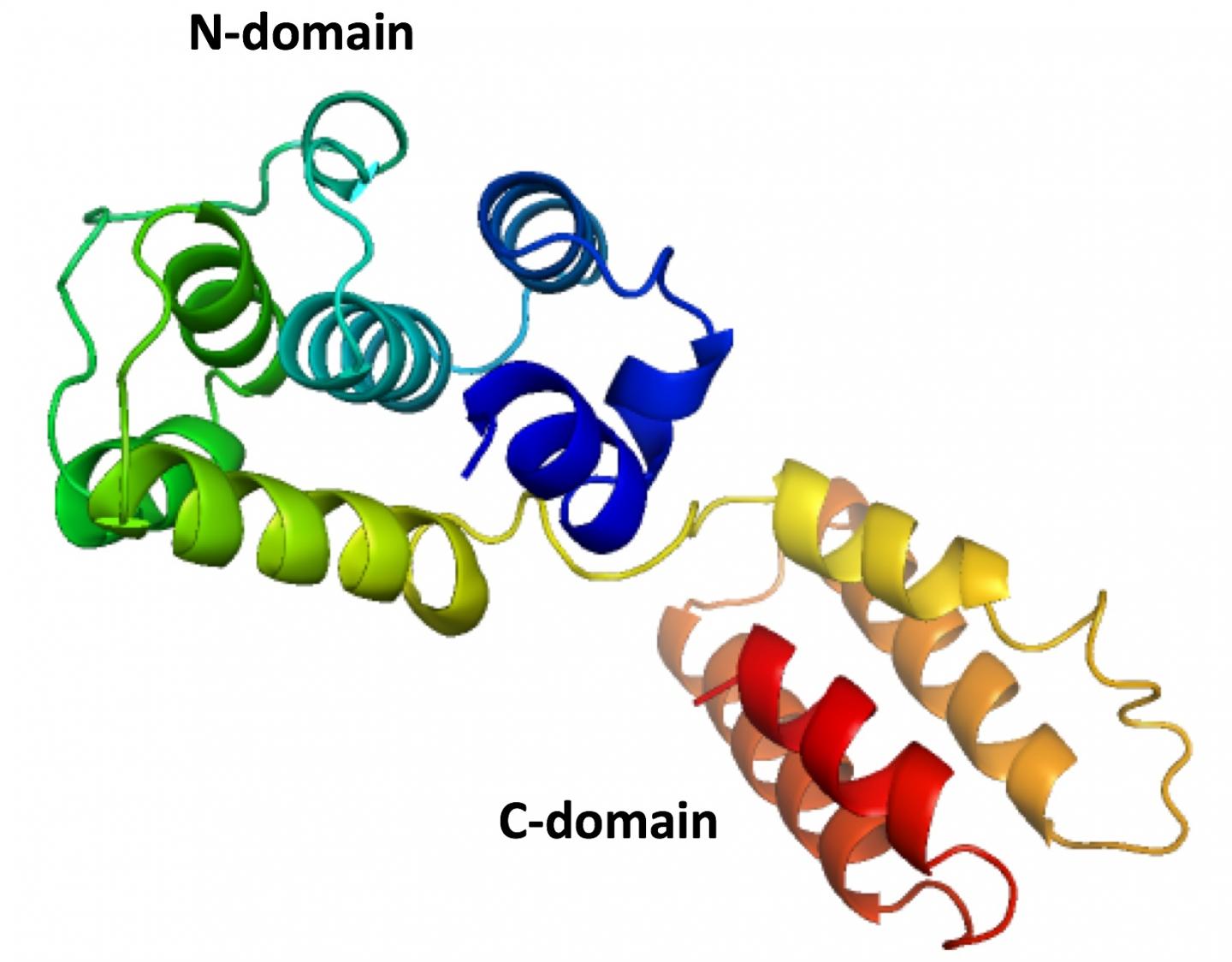
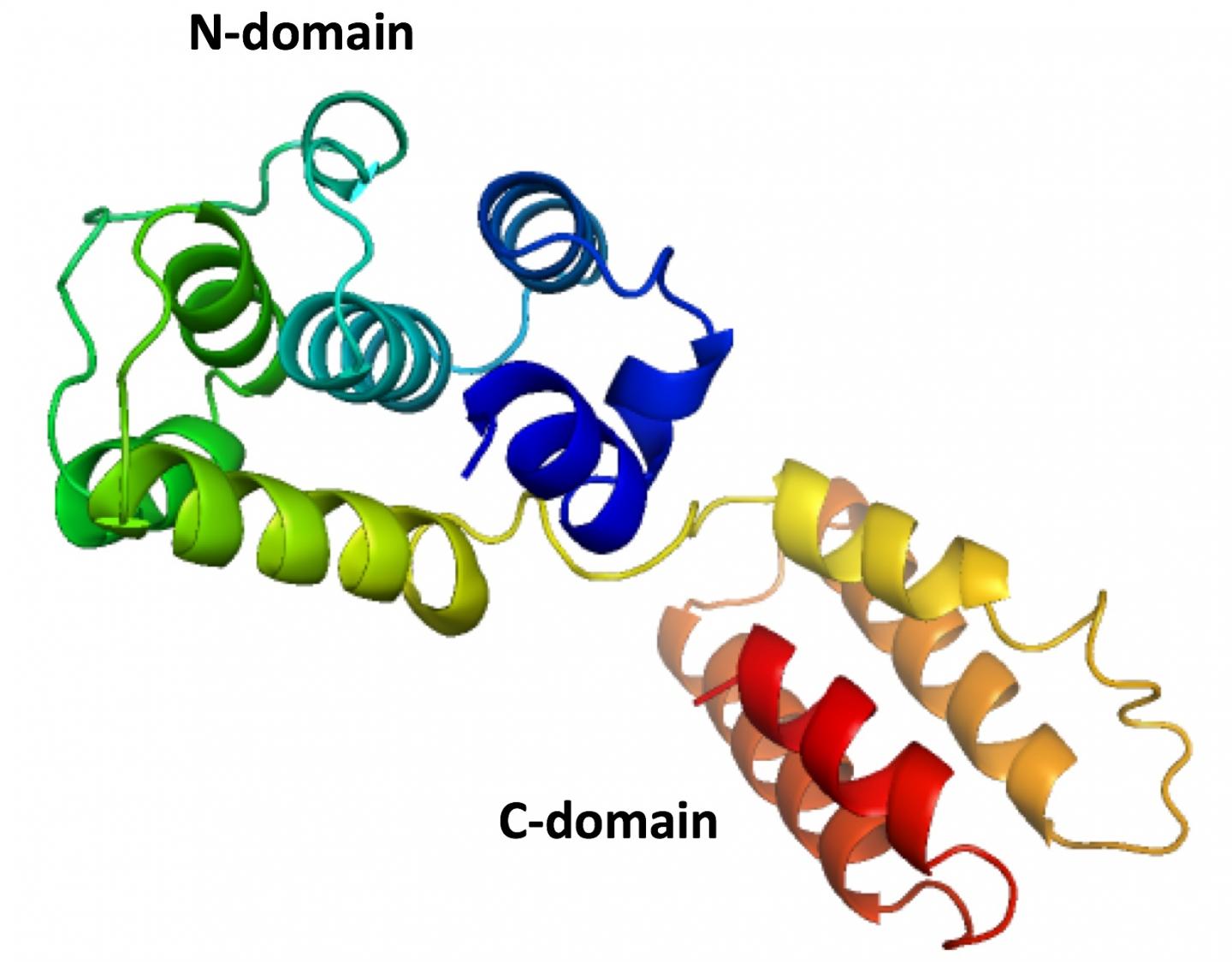
Tao's lab specializes in X-ray crystallography and other techniques to reveal the basic structures of molecules like the fragile matrix protein dubbed M1. Many attempts to define the complete structure have failed because the protein has two major sections: the N and C domains.
Because the thin strand of residues that holds the domains together falls apart so easily, researchers until now have only been able to capture N domain structures through crystallization.
M1 may be fragile on its own, but it becomes a tough shell when it binds with other M1 proteins to form a spherical or tubular matrix that encapsulates and protects the viral RNA. This matrix sits just inside a lipid bilayer, the outer envelope of the virus that incorporates long glycoproteins. These glycoproteins seek out and attach to target cells with their ectodomains, while their cytoplasmic tails serve as a bridge to the N domain of the matrix protein. The C domain points inward from the matrix and is a bridge to the viral load inside.
"M1 helps to support the shape," Tao said. "That is a major function for any matrix protein. The matrix also helps to incorporate the viral RNA. Presumably, if there's no matrix protein, you end up with an empty vesicle."
The elbow-shaped structure of the M1 found in the infectious salmon anemia virus has six tightly packed alpha helices that make up the N domain and closely resemble those found in strains of influenza A. On the other side of the flexible hinge, the "elbow," lies the C domain with four alpha helices. Both domains have hydrophobic cores to stabilize them around the hinge that can vary by as much as 40 degrees.
Tao said having the complete structure of the protein will help researchers determine how it and others like it polymerize into a protective shell and associate with the membrane. "Knowing the structure of only one domain, we could not understand how matrix proteins interacted with each other to form the shell," she said. "The interaction involves both domains."
She said the full structure could also provide clues for future flu antivirals. A treatment that addresses M1 would have to find and invade the virus-infected cells.
"But if there's a chemical that can interrupt the self-association of M1 proteins, it's going to be very useful," Tao said. "I don't think it matters which domain it binds, as long as it prevents the shell from forming. This could be a valid target."
###
Rice alumna Wenting Zhang, now a student at the Huazhong Agricultural University, Wuhan, China, is lead author of the paper. Co-authors from Rice are alumna Wenjie Zheng, postdoctoral fellow Yukimatsu Toh, graduate students Miguel Betancourt-Solis and Yanlin Fan, and James McNew, a professor of biochemistry and cell biology. Other co-authors are postdoctoral fellow Jiagang Tu and Jun Liu, an associate professor of pathology and laboratory medicine at the University of Texas Medical School at Houston; Vikram Vakharia, a professor at the University of Maryland Baltimore County; and Meilin Jin, a professor of preventative veterinary medicine at Huazhong Agricultural University. Tao is an associate professor of biochemistry and cell biology.
The research was supported by the Welch Foundation, the National Institutes of Health, the National Key Research and Development Program of China, the Hamill Foundation and the Kresge Science Initiative Endowment Fund at Rice University.
Read the abstract at http://www.pnas.org/content/early/2017/07/19/1701747114.abstract
This news release can be found online at http://news.rice.edu/2017/07/28/rice-u-scientists-reel-in-structure-of-salmon-virus/
Follow Rice News and Media Relations via Twitter @RiceUNews
Related materials:
Tao Laboratory: http://www.bioc.rice.edu/~ytao/
Wiess School of Natural Sciences: http://natsci.rice.edu
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation's top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,879 undergraduates and 2,861 graduate students, Rice's undergraduate student-to-faculty ratio is 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for happiest students and for lots of race/class interaction by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger's Personal Finance. To read "What they're saying about Rice," go to http://tinyurl.com/RiceUniversityoverview.
Media Contact
David Ruth
[email protected]
713-348-6327
@RiceUNews
http://news.rice.edu
Related Journal Article
http://dx.doi.org/10.1073/pnas.1701747114