NEW YORK – A collaboration between researchers at Columbia and the Icahn School of Medicine is embarking on a project that will generate comprehensive atlases of entire human brains and all their 180+ billion cells. This kind of data can help uncover how the structure and organization of the brain give rise to behavior, emotion and cognition, in sickness and in health.
Credit: Hillman Lab/Columbia’s Zuckerman Institute
NEW YORK – A collaboration between researchers at Columbia and the Icahn School of Medicine is embarking on a project that will generate comprehensive atlases of entire human brains and all their 180+ billion cells. This kind of data can help uncover how the structure and organization of the brain give rise to behavior, emotion and cognition, in sickness and in health.
Until now, the vast technical complexity and time needed for mapping the whole human brain has constrained cellular-level brain atlasing to much smaller animals or just portions of the human brain.
“Throughout the history of science, new tools have been behind some of the most dramatic advances,” said Elizabeth Hillman, PhD, a Herbert and Florence Irving Professor at Columbia’s Zuckerman Institute and leader of the project. “We are developing technologies that should make high-speed, large-scale imaging of tens or even hundreds of human brains a feasible prospect in the next five years. The unprecedented troves of data that we hope to produce should open the way to previously inaccessible knowledge about the human brain.”
To enable Dr. Hillman and her collaborators to undertake this ambitious project, the National Institutes of Health BRAIN Initiative recently awarded them a $9.1 million grant. The funding will be shared between Columbia University, the Icahn School of Medicine at Mount Sinai and Carnegie Mellon University. Since 2014, the BRAIN Initiative has invested over $2.4 billion in research funding to boost our understanding of how the brain works. The new project falls under the auspices of the BRAIN Initiative Cell Census Network, which was established in 2017 to encourage researchers to find ways to generate comprehensive brain-cell atlases.
“If successful, our microscope should be able to image an entire human brain with cellular detail in a matter of days,” said Dr. Hillman, who is also a professor of biomedical engineering and radiology at Columbia. “This data will be like Google Earth for the brain, enabling analysis of patterns and distributions of different types of human brain cells across vastly different length scales. To get a feel for the challenge, keep in mind that there are only eight billion people on earth but over 180 billion cells in the brain.”
The team doesn’t just want to count cells. A top priority is to develop a brain map charting the diversity of the many different kinds of cells that make up the brain.
“We know that the brain contains billions of neurons, but there are many different subtypes of neurons,” said Dr. Hillman. “How many there are, how they are organized and how they vary between different brain regions and different people is largely unknown.”
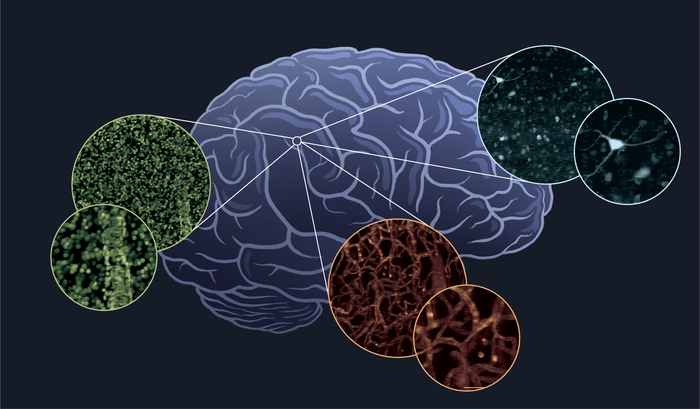
But the brain isn’t made only of neurons. Its meshwork includes other types of cells, among them a range of glial cells and cells making up the brain’s vasculature. All of these cell types are essential for normal brain function and could hold important clues about what goes wrong in disease.
“To make these datasets really useful, we have to find a way to capture as much information as possible as we scan the whole brain,” said Dr. Hillman, who has a track record of inventing new, powerful and fast microscope techniques.
For this brain-atlasing project, she is developing another new microscope technique. It’s called Human Brain Optimized Light Sheet (HOLiS) microscopy. The team chose the name to emphasize the importance of holistic imaging and analysis of the entire human brain of each individual.
The first step in the imaging process is to carefully cut the brain into 5-millimeter thick sections and process them to make them completely transparent. This almost magical feat is the specialty of co-Principal Investigator on the project, Zhuhao Wu, PhD, assistant professor at Mount Sinai’s Laboratory of Neural Systems, Structures and Genetics. Dr. Wu has optimized a method for the human brain clearing, which includes a step that can infuse each brain section with a range of fluorescent tags that make it possible to identify individual cells and their diverse properties based on their different colors.
Then comes the HOLiS microscope, which operates at lightning speed to generate massive, technicolor 3D images of each section. The technique works by projecting laser light into the tissue to create a sheet of light that illuminates a very thin tilted plane, while a fast camera captures an image of the same plane. By moving the brain section at constant speed, successive images of each plane can be stacked together to form a long 3D block. The tissue is then scanned back and forth to cover its whole volume before moving onto the next section.
“Attempting to image a whole human brain with existing conventional instruments would take years,” said Hillman. “We hope our HOLiS system will be able to image an entire brain in about a week.”
This kind of speed, added Hillman, will take whole-brain imaging from a one-off proof of concept to a technology capable of imaging hundreds of brains. “We suspect that every brain will be very different, so we need to be able to image a lot of brains to understand brain diversity across the lifespan, and to ultimately be able to explore a wide range of diseases and disorders.”
Another challenge remains however. The team expects each brain-atlasing run to generate some two petabytes of data, a massive amount. Collaborators at the Pittsburgh Supercomputing Center at Carnegie Mellon will help the team to convert these torrents of data into more manageable, searchable and user-friendly databases that can be analysed and compared. Contributing to this crucial aspect of the project with expertise in computer
science, machine vision, information theory and statistics are Carl Vondrick, PhD, and Cynthia Rush, PhD, from Columbia’s Data Science Institute as well as Luke Hammond, Director of the Zuckerman Institute’s Cellular Imaging Core.
Among others joining in the effort are Dr. Wu’s colleagues at the Icahn School of Medicine, including John F. Crary, MD, PhD, director of the school’s Neuropathology Brain Bank and an expert in human brain preservation and neuropathology. Alan Seifert, PhD, assistant professor at Mount Sinai’s Biomedical Engineering and Imaging Institute, will acquire detailed magnetic resonance images of the whole brain before it is cut. This will enable all of the data collected with HOLiS to be registered to current brain atlases and analyzed to compare cellular-level HOLIS data to MRI signal properties.The Icahn team also includes Bradley Delman, MD, professor of Radiology, and Patrick Hof, MD, professor of Neuroscience, who will contribute their special expertise in neuroradiology and neuroanatomical reading of human brain data.
Adding to the mix of talent on the project is Pavel Osten, MD, PhD, a pioneer in the field of whole-brain cellular imaging and now President, founder and Chief Scientific Officer of a new start-up company. Dr. Osten was instrumental in planning the project and will provide guidance and advice on the best ways to rapidly analyze HOLiS images to find all of the cells and to map information from HOLiS scans onto established anatomical atlases of the human brain.
“If we can streamline the process we can build a foundational database that enables analysis of the human brain like never before,” said Dr. Hillman. “Having this data should accelerate our efforts to understand what so often goes right in the human brain and what goes wrong in developmental, neurological and psychiatric disorders.
Award details are as follows:
“Cell type atlasing of whole human brains using HOLiS: an optimized pipeline for staining, clearing, imaging, and analysis” (1RF1MH128969-01)
Total Award: $9,121,879 over three years.
Method of Research
News article
Subject of Research
People




