Credit: Brown University
PROVIDENCE, R.I. [Brown University] — A team of Brown University researchers has made substantial progress in an effort to create a new type of molecular data storage system.
In a study published in Nature Communications, the team stored a variety of image files — a Picasso drawing, an image of the Egyptian god Anubis and others — in arrays of mixtures containing custom-synthesized small molecules. In all, the researchers stored more than 200 kilobytes of data, which they say is the most stored to date using small molecules. That’s not a lot of data compared to traditional means of storage, but it is significant progress in terms of small molecule storage, the researchers say.
“I think this is a substantial step forward,” said Jacob Rosenstein, an assistant professor in Brown’s School of Engineering and an author of the study. “The large numbers of unique small molecules, the amount of data we can store, and the reliability of the data readout shows real promise for scaling this up even further.”
As the data universe continues to expand, much work is being done to find new and more compact means of storage. By encoding data in molecules, it may be possible to store the equivalent of terabytes of data in just a few millimeters of space. Most research on molecular storage has focused on long-chain polymers like DNA, which are well known carriers of biological data. But there are potential advantages to using small molecules as opposed to long polymers. Small molecules are potentially easier and cheaper to produce than synthetic DNA, and in theory have an even higher storage capacity.
The Brown research team, supported by a U.S. Defense Advanced Research Projects Agency (DARPA) grant led by chemistry professor Brenda Rubenstein, has been working to find ways of making small-molecule data storage feasible and scalable.
To store data, the team uses small metal plates arrayed with 1,500 tiny spots less than a millimeter in diameter. Each spot contains a mixture of molecules. The presence or absence of different molecules in each mixture indicate the digital data. The number of bits in each mixture can be as large as the library of distinct molecules available for mixing. The data can then be read out using a mass spectrometer, which can identify the molecules present in each well.
In a paper published last year, the Brown team showed that they could store image files in the kilobyte range using some common metabolites, the molecules that organisms use to regulate metabolism. For this new work, the researchers were able to vastly expand the size of their library — and thereby the sizes of the files they could encode — by synthesizing their own molecules.
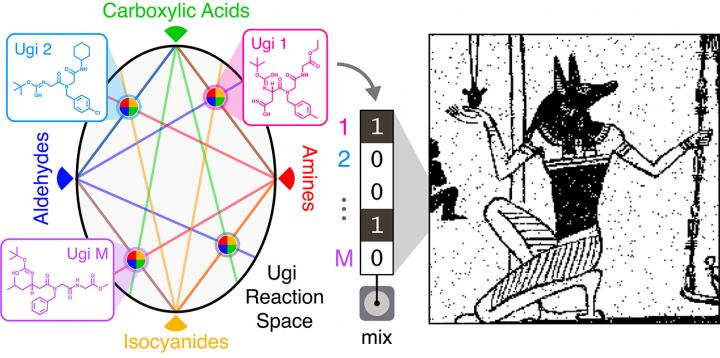
The team made their molecules using Ugi reactions — a technique often used in the pharmaceutical industry to quickly produce large numbers of different compounds. Ugi reactions combine four broad classes of reagents (an amine, an aldehyde or a ketone, a carboxylic acid, and an isocyanide) into one new molecule. By using different reagents from each class, the researchers could quickly produce a wide array of distinct molecules. For this work, the team used five different amines, five aldehydes, 12 carboxylic acids, and five isocyanides in different combinations to create 1,500 distinct compounds.
“The advantage here is the potential scalability of the library,” Rubenstein said. “We use just 27 different components to make a 1,500-molecule library in one day. That means we don’t have to go out and find 1,500 unique molecules.”
From there, the team used sub-libraries of compounds to encode their images. A 32-compound library was used to store a binary image of the Egyptian god Anubis. A 575-compound library was used to encode a 0.88-megapixel Picasso drawing of a violin.
The large number of molecules available for the chemical libraries also enabled the researchers to explore alternate encoding schemes that made the readout of data more robust. While mass spectrometry is highly precise, it’s not perfect. So as with any system used to store or transmit data, this system will need some form of error correction.
“The way we design the libraries and read out the data includes extra information that lets us correct some errors,” said Brown graduate student Chris Arcadia, first author of the paper. “That helped us streamline the experimental workflow and still get accuracy rates as high as 99 percent.”
There’s still more work to be done to bring this idea up to a useful scale, the researchers say. But the ability to create large chemical libraries and use them for encoding ever larger files suggests the approach can indeed be scaled up.
“We’re no longer limited by the size of our chemical library, which is really important,” Rosenstein said. “That’s the biggest step forward here. When we started this project a few years ago, we had some debates about whether something of this scale was even experimentally feasible. So it’s really encouraging that we’ve been able to do this.”
###
Other co-authors on the paper were Eamonn Kennedy, Joseph Geiser, Amanda Dombroski, Kady Oakley, Shui-Ling Chen, Leonard Sprague, Mustafa Ozmen, Jason Sello, Peter M. Weber, Sherief Reda, Christopher Rose and Eunsuk Kim. The work was funded by DARPA (W911NF-18-2-0031) and the National Science Foundation (1941344).
Media Contact
Kevin Stacey
[email protected]
401-863-3766
Original Source
https:/
Related Journal Article
http://dx.




