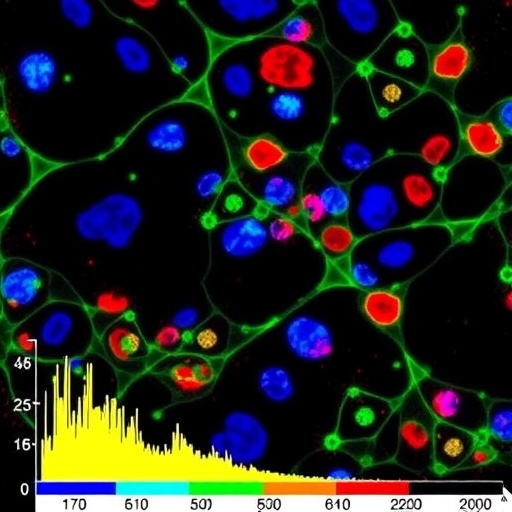
In a groundbreaking advancement that pushes the limits of cellular analysis, researchers have unveiled a novel technique capable of profiling intact proteoforms from thousands of individual cells extracted directly from the rat hippocampus. This achievement, enabled through an innovative mass spectrometry platform known as single-cell proteoform imaging mass spectrometry (scPiMS), heralds an entirely new era in neuroscientific and proteomic research. By capturing the full complexity of endogenous proteins at single-cell resolution, scientists gain an unprecedented window into the molecular heterogeneity of brain tissue, potentially transforming our understanding of neuronal diversity, brain function, and disease.
Traditional single-cell proteomics methods have long grappled with challenges related to sensitivity, throughput, and the ability to analyze whole proteins rather than peptide fragments. Although cutting-edge approaches have enabled identification of thousands of proteins at the single-cell level, they often rely on protein digestion, which obscures the proteoform landscape—distinct molecular variants arising from alternative splicing, post-translational modifications, and proteolytic processing. The scPiMS technology circumvents this limitation by directly extracting and analyzing intact proteins from individual cells, preserving vital information about their structural and functional diversity.
The researchers applied scPiMS to profile over 10,000 individual cells from the rat hippocampus, a brain region crucial for learning, memory, and spatial navigation. This scale of intact proteoform characterization from endogenous single cells is unprecedented and highlights the remarkable throughput and sensitivity of the platform. Utilizing an informatics pipeline specially designed for this complex data type, the team was able to classify primary brain cell populations—neurons, astrocytes, and microglia—based solely on their unique proteoform signatures, demonstrating the method’s capacity for high-resolution cellular phenotyping.
At the heart of the method lies the direct extraction of whole proteins from individual cells without chemical labeling or amplification. This strategy preserves the native state of proteins as they exist in cells, allowing detailed interrogation of proteoform diversity that arises from the myriad combinations of post-translational modifications and alternative isoforms. As a consequence, scPiMS provides a holistic snapshot of protein identity and function at the single-cell level, something that traditional bottom-up proteomic methods cannot fully capture.
The mass spectrometric workflow integrates high-resolution ion mobility separation alongside state-of-the-art mass analyzers, which together facilitate accurate mass measurement and isolation of intact proteoforms in a high-throughput manner. Crucially, this approach maintains both spatial resolution and quantitative capability, ensuring that the intrinsic proteomic signatures defining each cell type remain unaltered during analysis. The resulting data reveal rich heterogeneity within ostensibly homogeneous populations, uncovering subtle molecular nuances that may underlie diverse cellular states or functional specializations.
To translate the complex proteoform data into meaningful biological insights, the team engineered an informatics workflow optimized for scPiMS output. This analytical framework involves advanced algorithms for protein identification, proteoform assignment, and multivariate classification. By leveraging machine learning-based pattern recognition, the pipeline succeeded in robustly distinguishing neurons, astrocytes, and microglia according to their unique molecular fingerprints. The ability to assign cell identity at scale purely from proteoform profiles establishes a valuable tool for exploring cellular diversity without relying solely on transcriptomic or antibody-based markers.
This technological breakthrough holds profound implications for neuroscience, biomarker discovery, and systems biology at large. The rat hippocampus, characterized by complex cellular networks and dynamic functional states, provides an ideal proving ground for the capabilities of scPiMS. Through comprehensive proteoform profiling at single-cell resolution, researchers can now dissect the molecular underpinnings of neurological processes and disorders with a level of detail previously unattainable through proteomic avenues.
Moreover, the direct measurement of proteoforms addresses key biological questions related to protein modifications and isoform-specific functions. Since many neurodegenerative diseases, cognitive impairments, and psychiatric conditions are linked to aberrant protein processing and modifications, scPiMS may pave the way for identifying novel therapeutic targets by revealing disease-associated proteoform alterations within defined cell populations. The method also complements and extends single-cell transcriptomics by adding a critical proteomic dimension that reflects actual protein function and regulation in cells.
Importantly, the throughput of scPiMS allows practical application to thousands of cells, enabling statistically robust analyses of heterogeneous tissues. This scaling overcomes one of the fundamental bottlenecks of intact proteoform mass spectrometry, which traditionally required extensive sample preparation or yielded data from only a handful of cells. By demonstrating that endogenous whole proteins can be profiled en masse directly from single cells, the investigators establish a new paradigm for future efforts in proteomic single-cell atlasing across diverse tissues and organisms.
The wealth of proteoform information obtained by this technique also opens avenues for integrative multi-omics approaches. When combined with spatial transcriptomics and imaging modalities, scPiMS-derived data can illuminate the interplay between gene expression, protein regulation, and cellular phenotype in situ. Such comprehensive molecular portraits will accelerate efforts to decode complex biological systems and better understand how cellular heterogeneity shapes physiological and pathological states.
Though the foundation laid by this study is promising, there remain challenges to be addressed, including deepening proteome coverage, further improving sensitivity to detect low-abundance proteoforms, and adapting the technology to human tissues and clinical samples. Continued refinement of informatics workflows and integration with complementary data types will enhance the biological interpretability of these rich datasets. Nonetheless, the successful implementation of scPiMS marks a pivotal step forward in the field of single-cell proteomics.
The authors of this work envision that scPiMS technology will soon empower researchers to unravel the detailed proteomic landscapes of complex tissues with unparalleled fidelity. As the method matures, it may prove invaluable for elucidating cellular interactions in brain circuits, monitoring disease progression at the molecular level, and guiding precision medicine strategies tailored to cellular proteoform profiles. The ability to resolve proteoform diversity at scale from endogenous single cells promises to enrich our molecular vocabulary for understanding life’s complexity.
In summary, the advent of single-cell proteoform imaging mass spectrometry represents a quantum leap in proteomic analysis. By capturing intact proteins across thousands of individual cells from the rat hippocampus, this approach moves beyond fragmented peptide data to reveal the full proteoform panorama underpinning cellular identity and function. This technological breakthrough lays the groundwork for exciting biological discoveries and highlights the transformative potential of integrating proteomics deep into the single-cell realm.
As the scientific community embraces scPiMS, future applications could span myriad areas including developmental biology, neurodegeneration, immunology, and cancer research. The capacity to define cell types and states through their comprehensive proteoform signatures will augment existing single-cell technologies and offer new insights inaccessible by other means. This innovative fusion of mass spectrometry and single-cell resolution promises to illuminate the proteomic intricacies of cellular life and inspire a wave of discoveries illuminating the brain and beyond.
—
Subject of Research: Single-cell proteoform profiling of endogenous cells from the rat hippocampus using imaging mass spectrometry
Article Title: Proteoform profiling of endogenous single cells from rat hippocampus at scale
Article References:
Su, P., Hollas, M.A.R., Pla, I. et al. Proteoform profiling of endogenous single cells from rat hippocampus at scale.
Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02669-x
Image Credits: AI Generated
Tags: alternative splicing effectsbrain function researchcellular analysis innovationsintact protein profilingmass spectrometry techniquesmolecular heterogeneity in brain tissueneuronal diversity analysispost-translational modificationsproteomic advancementsrat hippocampus proteoformsscPiMS technologysingle-cell proteomics