Credit: Jason Cheng
Drug resistance is a major obstacle to effective treatment for patients with cancer and leukemia. Epigenetic modifying drugs have been proven effective for some patients with hematologic malignancies, such as myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). Unfortunately, most patients treated with epigenetic modifying drugs develop resistance, resulting in therapeutic failure and eventually, patient death. The mechanisms underlying the selectivity and resistance of epigenetic modifying drugs remain largely elusive.
A research team based at the University of Chicago has begun to unravel the role of RNA epigenetics and chromatin structure in regulation of 5-azacytidine (5-AZA), a well-known DNA hypomethylating agent in MDS and AML. The finding may lead to novel strategies, as well as guidance from clinical biomarkers that could help predict and reduce the risk of drug resistance, a major obstacle in leukemia treatment.
"This is the first study to demonstrate that RNA cytosine methylation and methyltransferases mediate cell lineage-associated drug-responsive chromatin structures in MDS and AML," said the study's lead author, Jason Cheng, MD, PhD, assistant professor of pathology at the University of Chicago.
"This is a new area," he added. "Although a large number of RNA modifications have been identified in the past, the function and the clinical potential of those RNA modifications and their effects on gene regulation and chromatin organization remain largely unexplored."
RNA and DNA are composed of four nucleobases-adenine (A), cytosine (C), guanine (G), uracil (U) in RNA and thymine (T) in DNA. DNA associates with other proteins, such as histones, and RNA to form a large macromolecular complex called chromatin.
In human cells, chromatin is organized into distinct structures with functional domains that regulate gene expression and stem cell development. Chemical modifications of histones, DNA and RNA can change chromatin structures.
Methylation, one such chemical modification, adds a single carbon and three hydrogen atoms (called a methyl group) to other molecules, such as DNA, RNA and proteins. In mammalian cells DNA methylation occurs at cytosine residues of cytosine-phosphate-guanine (CpG) sequences, which leads to repression of gene expression. However, little is known about the function and the clinical significance of RNA cytosine methylation (RNA:m5C) and their modifying enzymes, such as RNA cytosine methyltransferases (RCMTs), that add the methyl group to cytosines of RNA.
The research team was particularly interested in the role of RNA:m5C and RCMTs in regulation of cell lineage-associated chromatin structures and drug response/resistance in MDS and AML. They found a significant increase in RNA:m5C and RCMTs in 5-AZA-resistant leukemia cells compared to that in 5-AZA-sensitive leukemia cells.
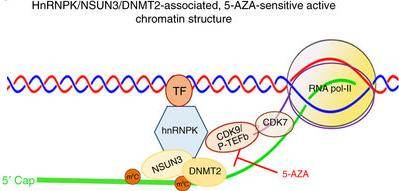
They show that 5-AZA-sensitive and 5-AZA-resistant leukemia cells have distinctly different chromatin structures that are associated with RNA:m5C and RCMTs at nascent RNA, as schematically illustrated.
Specifically, in 5-AZA-sensitive leukemia cells, RCMTs–namely NSUN3 and DNMT2–bind hnRNPK, a conserved RNA-binding protein, to recruit active RNA polymerase II (RNA-pol-II) to form an active chromatin structure at nascent RNA.
In contrast, a different RCMT, called NSUN1, interacts with BRD4, a histone modifier, to recruit RNA-pol-II to form a different active chromatin structure at nascent 5-AZA-resistant leukemia cells. This NSUN1/BRD4-mediated chromatin structure renders these leukemia cells insensitive to 5-AZA, but hypersensitive to the BRD4 inhibitor, JQ1, and to the downregulation of NSUN1 by siRNAs.
By applying new technologies–including mass spectrometric analysis and proximity ligation rolling circle amplification confocal microscopy–to a small size of clinical MDS/AML specimens (n=18), the researchers demonstrated a significant increase in RNA:m5C and RCMTs and the NSUN1/BRD4-associated chromatin structure in 5-AZA-resistant vs. 5-AZA-sensitive clinical MDS/AML specimens.
"With the advent of modern molecular and imaging technologies, functional genomics will become a central platform to elucidate the function of genes, signal pathways and genetic networks, to determine the pathogenetic roles of gene mutations and provide powerful tools for better prognosis and more effective treatment for cancer/leukemia patients," Cheng said.
"We are moving towards functional genomics through exploring the potential of using RNA epigenetics and chromatin structures as diagnostic tools and potential therapeutic targets in MDS/AML patients," he added.
Additional studies are needed to identify the genetic alterations underlying the RNA epigenetics-mediated drug-responsive chromatin structures in MDS and AML. A large clinical study is necessary to determine the full potential and the limitation of such RNA epigenetics and chromatin structure-driven strategies in the future.
###
This study was supported by the Cancer Research Foundation, the American Cancer Society, a support grant to the University of Chicago Medicine Comprehensive Cancer Center, Swim Across America and the University of Chicago. Additional authors were Li Chen, Yuan Li, Adam Cloe, Ming Yue, Jiangbo Wei, John Anastasi, Richard Larson, Chuan He, Michelle Le Beau and James Vardiman from the University of Chicago; Kenneth Watanabe from Emory University; Jamile Shammo from Rush University Medical Center, and Qingxi Shen from the University of Nevada.
Media Contact
John Easton
[email protected]
773-795-5225
@UChicagoMed
http://www.uchospitals.edu
Related Journal Article
http://dx.doi.org/10.1038/s41467-018-03513-4