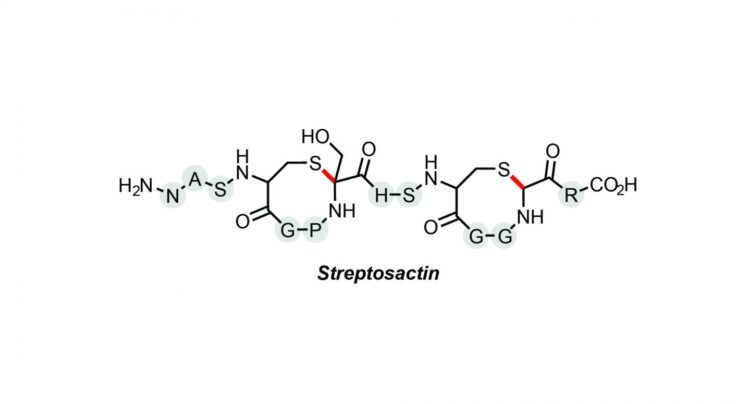
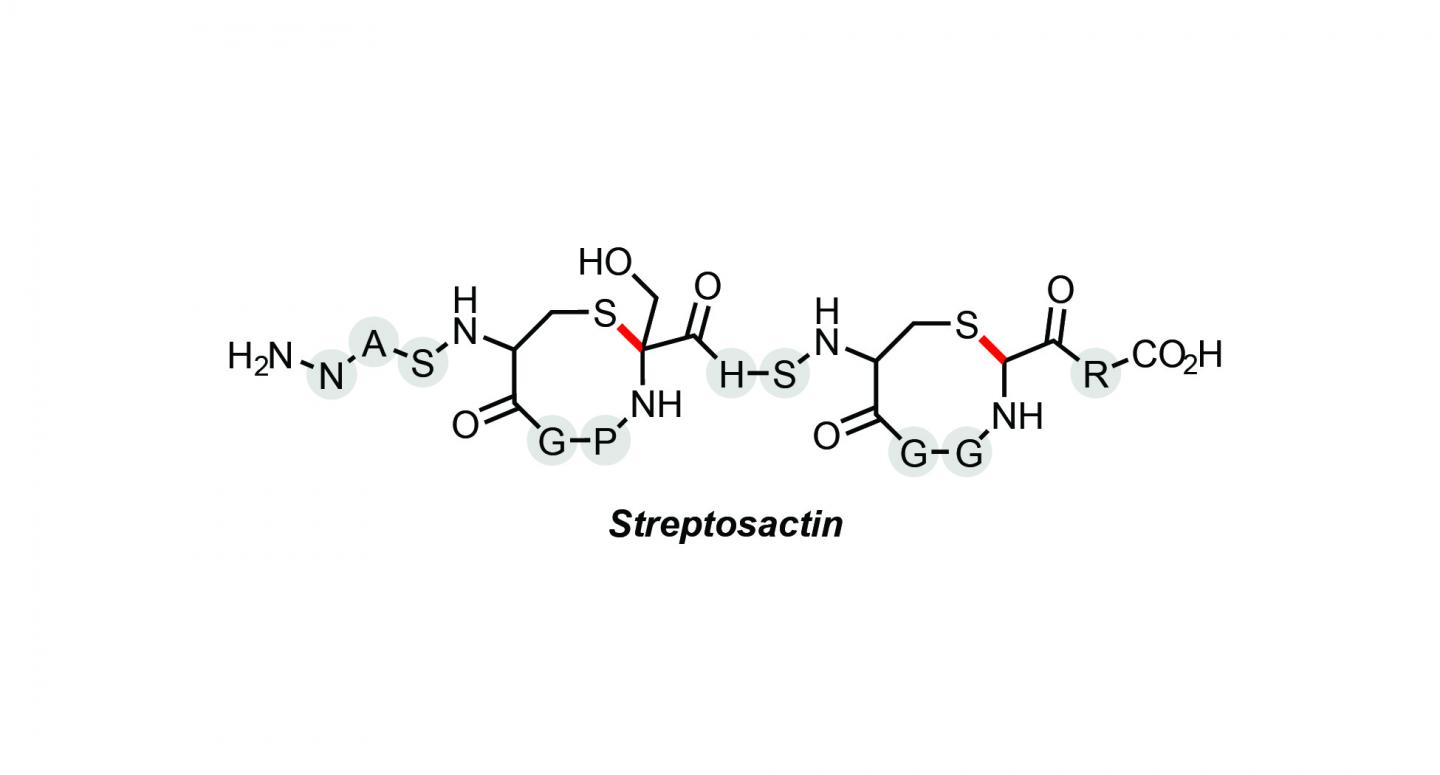
Streptosactin named the first small molecule in the human microbiome to exhibit fratricidal activity.
Credit: Figure courtesy of Leah Bushin.
A new bacterial molecule with the unsavory tendency to track down and kill others of its own kind has been discovered in the human microbiome by researchers at Princeton’s Department of Chemistry. Named Streptosactin, it is the first small molecule found to exhibit fratricidal activity.
The discovery by the Seyedsayamdost lab is detailed in the Journal of the American Chemical Society (JACS).
The research describes a veritable needle-in-a-haystack hunt in which streptosactin was located “at the edge of detection.” Its production is so minimal and so difficult to track that researchers achieved the results only through a complex interplay of factors.
They used a clever bioinformatic search strategy developed a few years ago by Leah Bushin, a sixth-year graduate student in the Seyedsayamdost lab. This “genome-first” approach allowed them to screen molecules for two key characteristics: community behavior (thus uncovering the fratricide), and structural or topological novelty.
The researchers then used advanced mass spectrometry tactics to separate out the “signal from noise,” manipulating and concentrating the culture extracts over a thousand-fold before locating the sought-after compound after months of searching.
The Cain-and-Abel behavior has been found in another bacterium where enzymes or large proteins provide the fratricidal activity – but never before in a small molecule. The purpose of the behavior is not fully understood. Researchers believe fratricide could contribute to evolutionary fitness by enhancing the genetic makeup of the sibling population.
“It’s crazy, and I was surprised by it, too. This bioactivity is something we would never have predicted,” said Mohammad Seyedsayamdost, associate professor of chemistry and associated faculty in molecular biology. “But that’s how microbes live – there is no morality. It’s just raw survival.
“Fratricide is good for the fitness or strength of this species,” he added. “It’s one way in which this bacterium generates a diverse genetic makeup, so it can increase the chances of surviving any challenge or condition it encounters.”
An assist from genome sequencing
The lab’s research opens a new window onto the vast, largely unexplored microbiome, that aggregate of microorganisms that lives in the mammalian body and likely plays a significant role in the health of all mammals. So uncharted is the microbiome that it has been described as a frontier every bit as exotic as the surface of the moon or the bottom of the ocean.
Natural products chemists like those in the Princeton lab try to discover and understand the molecules that microbes use to interact with their environments. Those interactions allow them to work together, to acquire food material, to compete, even to kill each other. Scientists then use this information to generate drug leads for antibiotics and antivirals; in fact, some 70% of all antibiotics we use clinically come from these sources.
“Natural product research can be really frustrating because luck plays a significant role. In a way it’s a little like fishing,” said Bushin, the lead author on the paper Discovery and Biosynthesis of Streptosactin, a Sactipeptide with an Alternative Topology Encoded by Commensal Bacteria in the Human Microbiome, which appears in JACS this week.
“After months of searching, you think you have a hit and then you go through the process of characterizing it – is it new? Is it known? If it isn’t, you have to start from square one. But when you do find something new like streptosactin, it’s this rush of adrenaline that’s unbelievable. That’s why you go through the months of hard work. As a scientist, it’s definitely what gets me into the lab every day.”
Mass Spectrometry Delivers
Even with a new search approach and an extremely sensitive instrument, streptosactin was difficult to identify from live cultures; its concentration was measured in the picomolar range.
After bioinformatic prediction and characterization of the biosynthetic enzymes, Brett Covington, a postdoctoral researcher in the Seyedsayamdost lab, managed to pinpoint the molecule in bacterial culture extracts. He cultured the organism “in a medium that it likes,” and used clever mass spectrometry tactics to locate the chemical fingerprints he was looking for.
“The problem we had with streptosactin is that, in lab cultures, it’s not produced well at all. We have a really sensitive instrument and still, it’s barely detectible over the noise,” said Covington. “So for months I was looking at things, asking could they be noise or could they be real?
“It’s a roller coaster with natural products chemistry,” Covington added. “If we had a little bit worse of an instrument, we wouldn’t have found it. If we didn’t concentrate it as well, we wouldn’t have found it. If we didn’t have the right types of methods that we were using, we wouldn’t have seen it. You had to know exactly what you were looking for and you had to look for a really long time.”
Bushin said, “I truly believe Brett is one of only a few people in the world who could have completed this task.”
The properties and behavior of streptosactin have yet to be puzzled through: what is its role in the microbiome? Why the fratricidal behavior? And what other discoveries remain from the more than 600 natural products predicted by Bushin’s search algorithm? These will form the basis of future research projects spearheaded by the Seyedsayamdost lab.
“There are multiple these projects that can be supported by this work,” said Seyedsayamdost.
###
Read the full paper here: https:/
The research reported in the paper, “Discovery and Biosynthesis of Streptosactin, a Sactipeptide with an Alternative Topology Encoded by Commensal Bacteria in the Human Microbiome,” was supported by funding from the National Science Foundation (NSF GRFP Award and NSF CAREER Award), and the Burroughs Wellcome Fund PATH Investigator Award.
Media Contact
Wendy Plump
[email protected]
Original Source
http://pubs.