“Effectively integrating these rich resources with GWAS results will continue to help prioritize causative inherited genetic variants and improve our molecular understanding of disease etiology […]”
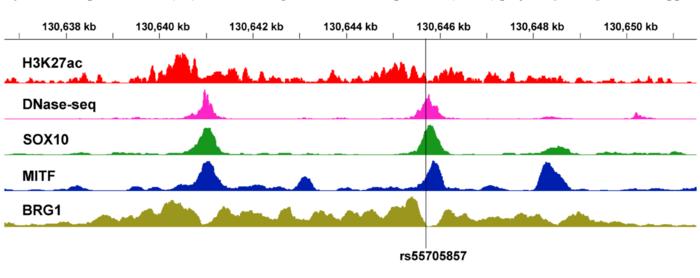
Credit: 2023 Song and Manjunath.
“Effectively integrating these rich resources with GWAS results will continue to help prioritize causative inherited genetic variants and improve our molecular understanding of disease etiology […]”
BUFFALO, NY- November 20, 2023 – A new editorial paper was published in Oncotarget’s Volume 14 on August 30, 2023, entitled, “Predicting the molecular functions of regulatory genetic variants associated with cancer.”
Some of inherited human genetic variation can contribute to important phenotypic diversity, such as the varying degrees of individual susceptibility to developing certain health conditions and individual response to therapeutic interventions. To date, over 490,000 genotype-phenotype associations have been discovered through large-scale genome-wide association studies (GWAS); however, molecular functions of most of these discovered GWAS variants remain unknown.
In their recent editorial, researchers Jun S. Song and Mohith Manjunath from the University of Illinois at Urbana-Champaign discuss computational methods of genetic analysis using expression quantitative trait loci, frameworks for predicting regulatory genetic functions, application to transcription factors involved in cancer development and progression, and future implications for their methods in cancer research and precision medicine.
“There are several technical challenges hindering our understanding […].”
First, the effect size of a typical genetic variant, as measured in terms of the odds ratio of genotype occurrence in case versus control populations, is very small, suggesting that macroscopic systems-level phenotypic differences modulated by each variant may also be small and difficult to detect. Next, most reported variants reside in non-protein-coding regions of the human genome, indicating that they are likely affecting the regulation of some unknown target genes’ expression. Finally, the discovered variants may not be functional themselves, but be merely in genetic linkage disequilibrium with other functional variants.
“A promising approach to address these challenges is to integrate genomic, epigenomic, transcriptomic and machine learning methods to identify functional genetic variants and characterize their mode of action in regulating target genes.”
Read the full paper: DOI: https://doi.org/10.18632/oncotarget.28451
Correspondence to: Jun S. Song
Email: [email protected]
Keywords: genome-wide association studies, cancer risk, regulatory variants, functional genomics
About Oncotarget: Oncotarget (a primarily oncology-focused, peer-reviewed, open access journal) aims to maximize research impact through insightful peer-review; eliminate borders between specialties by linking different fields of oncology, cancer research and biomedical sciences; and foster application of basic and clinical science.
To learn more about Oncotarget, visit Oncotarget.com and connect with us on social media:
- X, formerly known as Twitter
- YouTube
- LabTube
- Soundcloud
Sign up for free Altmetric alerts about this article: https://oncotarget.altmetric.com/details/email_updates?id=10.18632%2Foncotarget.28451
Click here to subscribe to Oncotarget publication updates.
For media inquiries, please contact: [email protected].
Oncotarget Journal Office
6666 East Quaker Str., Suite 1A
Orchard Park, NY 14127
Phone: 1-800-922-0957 (option 2)
###
Journal
Oncotarget
DOI
10.18632/oncotarget.28451
Method of Research
Data/statistical analysis
Subject of Research
Cells
Article Title
Predicting the molecular functions of regulatory genetic variants associated with cancer
Article Publication Date
30-Aug-2023




