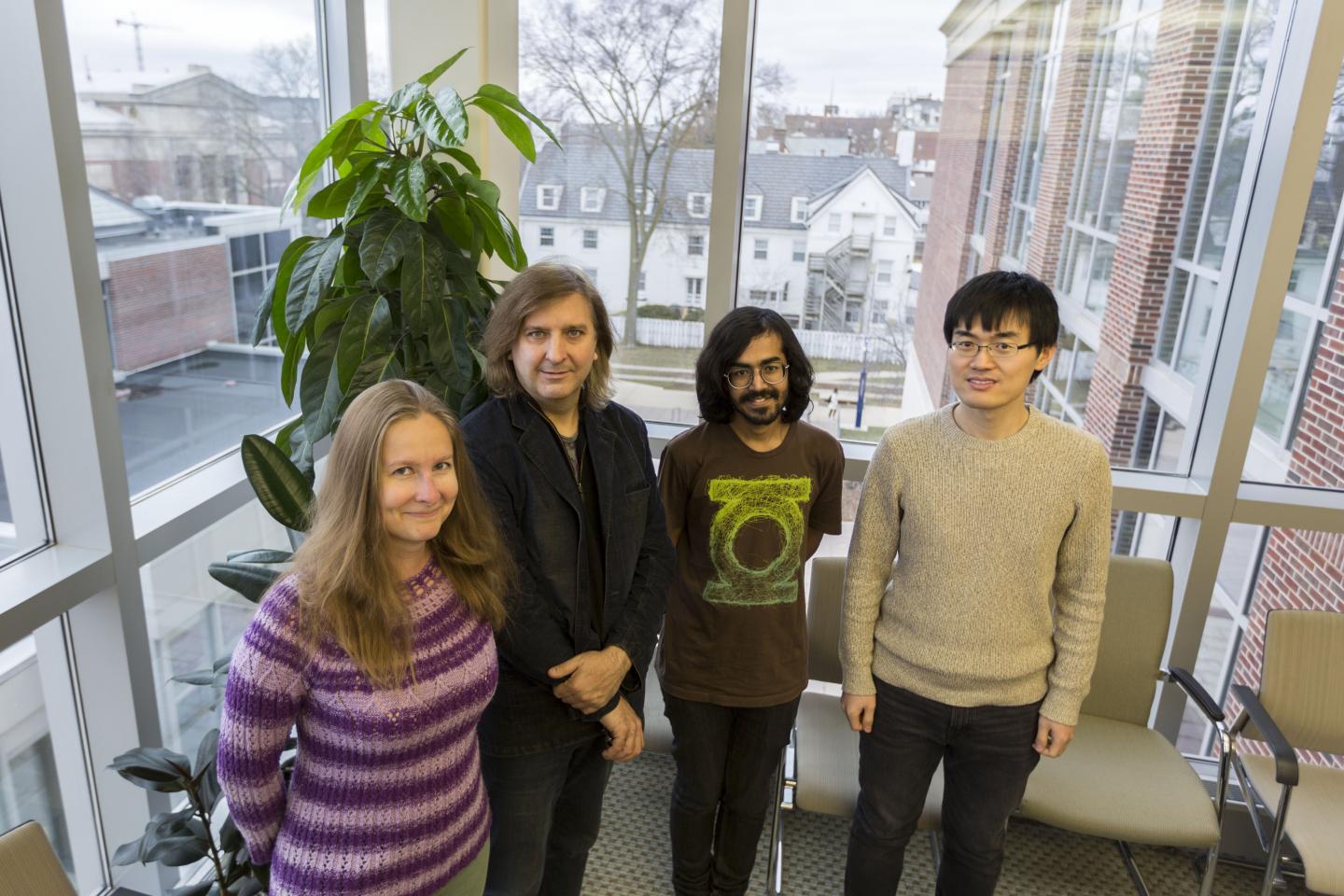
Credit: Julia Pollack, University of Illinois.
The human gut consists of a complex community of microbes that consume and secrete hundreds of small molecules–a phenomenon called cross-feeding. However, it is challenging to study these processes experimentally. A new study, published in Nature Communications, uses models to predict cross-feeding interactions between microbial species in the gut. Predictions from such computational methods could eventually help doctors get a more complete understanding of gut health.
The microbial community, or microbiome, of the gut is known to influence human health. Previous studies have focused on determining the types of microbes that are present. Unfortunately, this information is not enough to understand the microbiome.
“The gut environment is shaped by small molecules known as metabolites, which are excreted by the microbial community,” said Sergei Maslov (BCXT/CABBI), a professor of bioengineering and Bliss faculty scholar. “Although it is possible to measure these metabolites experimentally, it is cumbersome and expensive.”
The researchers had previously published a study where they used experimental data from other studies to model the fate of metabolites as they pass through the gut microbiome. In the new study, they have used the same model to predict new microbial processes that have not been determined before.
“What we eat passes into our gut, and there is a cascade of microbes which release metabolites,” said Akshit Goyal, a postdoctoral fellow at MIT and a collaborator of the Maslov lab. “Biologists have measured these molecules in human stools, we have shown that you can use computational models to predict the levels of some.”
Measuring every metabolite and trying to understand which microbe might be releasing it can be challenging. “There is a large universe of possible cross-feeding interactions. Using this model, we can aid experiments by predicting which ones are more likely to occur in the gut,” Goyal said.
The model was also supported by genomic annotations, which explain which microbial genes are responsible for processing the metabolites. “We are confident of our modelling predictions because we also checked whether the microbes contain the genes necessary for carrying out the associated reactions. About 65% of our predictions were supported by this information,” said Veronika Dubinkina, a PhD student in bioengineering.
The researchers are now working to improve the model by including more experimental data. “Different people have different strains of gut microbes. Although these different strains have many genes in common, they differ in their capabilities,” Dubinkina said. “We need to collect more data from patients to understand how different microbial communities behave in different hosts.”
“We are also interested in determining how fast the microbes consume and secrete the metabolites,” said Tong Wang, a PhD student in physics. “Currently the model assumes that all the microbes consume metabolites at the same rate. In reality, the rates are different and we need to understand them to capture the metabolite composition in the gut.”
###
The study “Ecology-guided prediction of cross-feeding interactions
in the human gut microbiome” can be found at https:/
Funding: Akshit Garg was supported by the Gordon and Betty Moore Foundation as a Physics of Living Systems Fellow.
Media Contact
Ananya Sen
[email protected]
Original Source
https:/
Related Journal Article
http://dx.