Credit: Penn State University
New, license-free DNA ladders will allow researchers to estimate the size of fragments of DNA for a fraction of the cost of currently available methods. A research team of undergraduate students led by Penn State Professor of Biochemistry and Molecular Biology Song Tan and former undergraduate student Ryan C. Henrici developed two plasmids — a circular form of DNA — that can be cut by DNA scissors known as restriction enzymes to create the DNA ladders. The ladders can be used to estimate the size of DNA fragments between about 50 and 5,000 base pairs in length. A paper describing the research appears online May 26, 2017 in the journal Scientific Reports.
"DNA ladders, also known as DNA molecular weight markers, are among the most commonly used reagents in molecular biology research," said Tan. "They are used in any application that requires gel electrophoresis — a technique that separates fragments of DNA by their size. We would like to offer these plasmids to the research community as a means to produce high quality DNA molecular weight markers at a low cost."
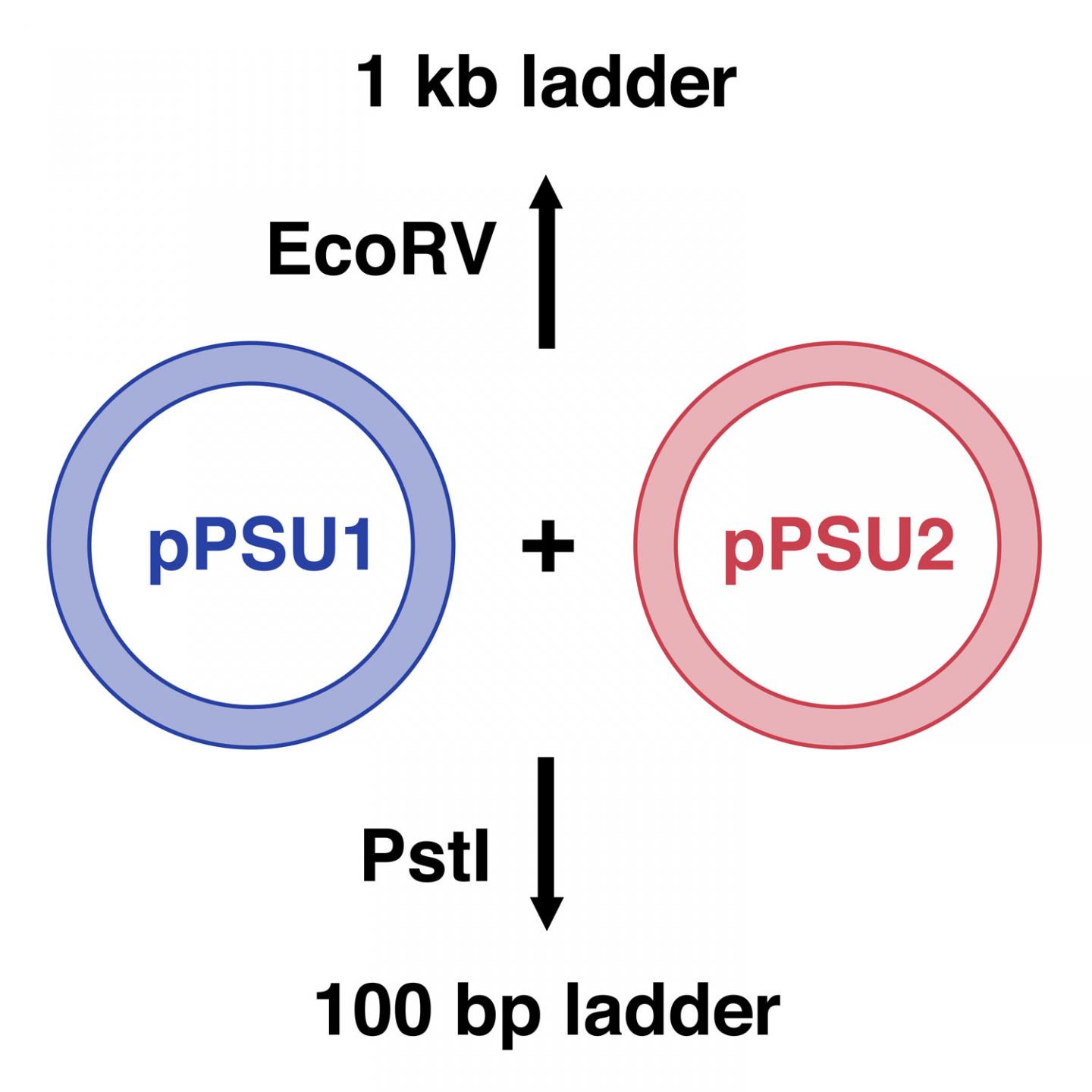
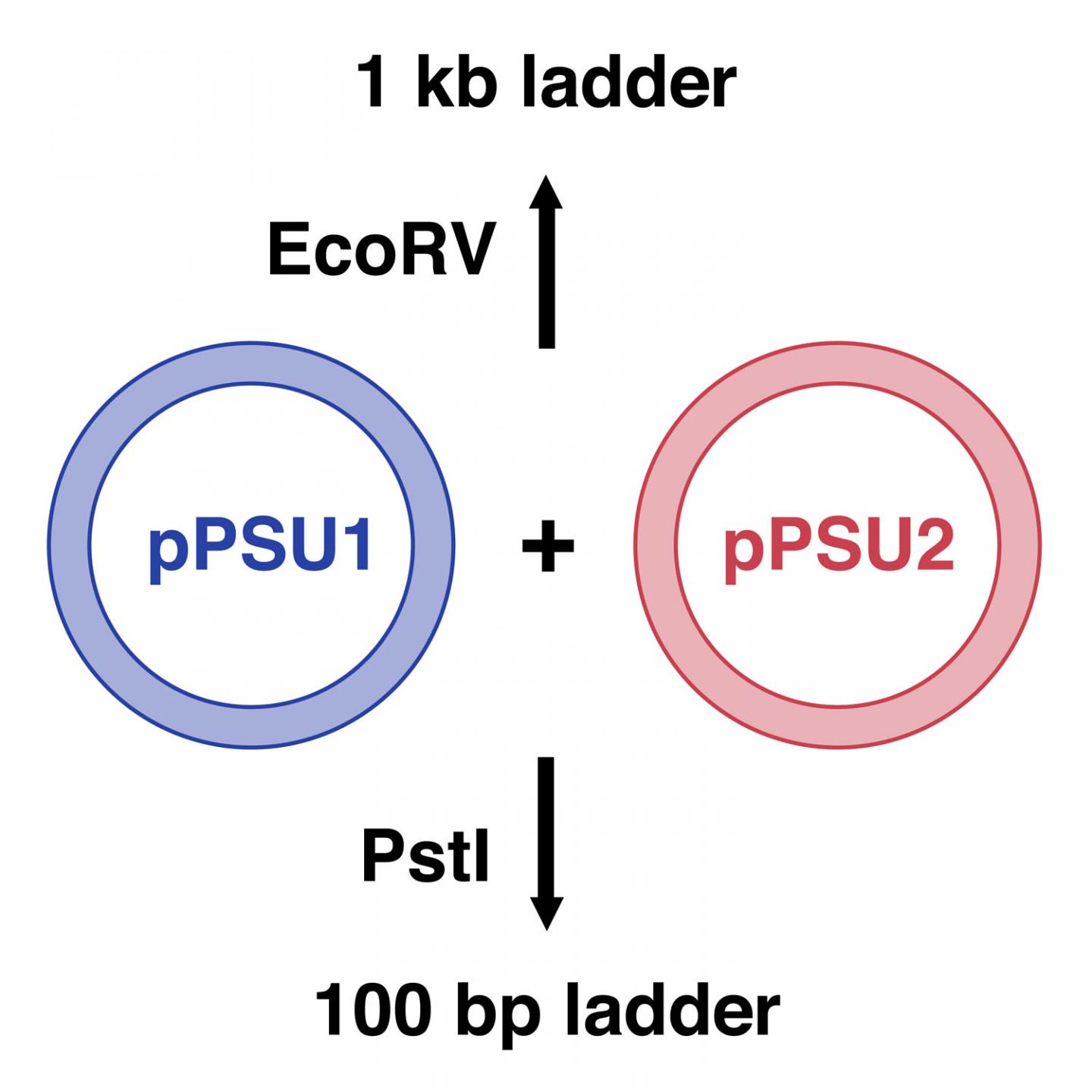
The research team created two plasmids, pPSU1 and pPSU2, that together produce DNA ladders in increments of either 100 or 1,000 base pairs, depending on which restriction enzyme is used. Researchers can easily produce in their own laboratories enough of the two ladders for 1,000 uses for under $10. In contrast, commercially available DNA ladders cost between $250 and $500 for the same amount. Additionally, unlike many currently available DNA ladders, the 100-base-pair ladders work appropriately on both agarose and polyacrylamide gels, two types commonly used in molecular biology.
"We are also excited about the possibility that the pPSU plasmids may be used around the world to further research and enhance science education in classroom laboratories," said Henrici. "This technology produces DNA ladders at less than a penny per use, a fraction of the cost of using commercially available DNA ladders."
The pPSU1 and pPSU2 plasmids used to produce the Penn State DNA ladders will be available without licensing restrictions to nonprofit academic users through the Addgene and DNASU plasmid repositories.
###
In addition to Tan and Henrici, the research team includes Turner J. Pecen and James L. Johnston, undergraduate students in the Schreyer Honors College at Penn State at the time the research was performed. The research is supported by the U.S. National Institutes of Health – National Institute of General Medical and the Penn State Eberly College of Science.
CONTACT:
Song Tan: [email protected], 814-865-3355
Barbara K. Kennedy (PIO): [email protected], 814-863-4682
IMAGES:
https://psu.box.com/v/Tan5-2017
CAPTIONS:
Figure 1: Schematic of pPSU1 and pPSU2 plasmids. The 1000-base-pair (1 kb) ladder is made by cutting the two plasmids with the restriction enzyme, EcoRV. The 100-base-pair (bp) ladder is made by cutting the plasmids with PstI.
Figure 2: Gel electrophoresis images of the two Penn State DNA ladders created by cutting the pPSU1 and pPSU2 plasmids with restriction enzymes. The 100-base-pair (bp) ladder is designed to be used with both agarose and acrylamide gels.
Media Contact
Barbara K. Kennedy
[email protected]
814-863-4682
@penn_state
http://live.psu.edu
############
Story Source: Materials provided by Scienmag