In a groundbreaking study published in Science Signaling, researchers from the University of Tokyo, led by Keigo Morita and Shinya Kuroda, have uncovered a complex interplay between structural stability and temporal coordination within the metabolic networks of the liver during starvation. This pioneering work sheds light on previously uncharted temporal vulnerabilities in obese mice, despite the metabolic network’s apparent structural robustness. Their novel approach, integrating multi-omics data with dynamic temporal analysis, opens new avenues for understanding metabolism’s intricate regulation under stress conditions and disease progression.
Metabolism is a finely tuned system essential to maintaining an organism’s homeostasis—the stable internal environment necessary for survival. Central to this system is the liver, which orchestrates numerous metabolic pathways, regulating both the quantity and timing of molecular activities. Starvation presents a critical physiological challenge, demanding precise adaptation at the cellular and molecular levels to conserve energy and sustain vital functions. While the liver’s responses to starvation have been extensively studied, few investigations have integrated the crucial temporal dimension that governs molecular interactions over time.
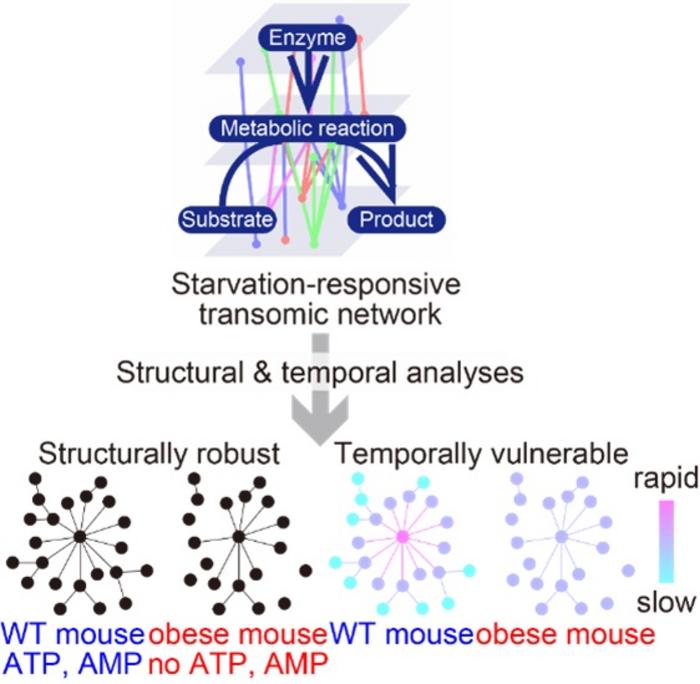
Morita, Kuroda, and their team focused their research on the starvation-responsive transomic network—a comprehensive map capturing interactions across diverse molecular layers, including metabolites, enzymes, and gene expression regulators. This network exemplifies the complexity of intracellular coordination required during metabolic stress. Particularly, they contrasted the liver networks of healthy versus obese mice, which exhibit distinct metabolic disturbances. Their research revealed that although the overall architecture of the metabolic network remained structurally intact in obesity, the temporal coordination—the precise timing with which hub molecules respond to starvation—was markedly compromised.
Fundamental to their discovery was the identification of “hub molecules,” key regulators exerting influence over many metabolic reactions. In healthy mice, hub molecules such as ATP and AMP, critical energy-sensing nucleotides, played a pivotal role in rapidly initiating adaptive responses to energy scarcity. Surprisingly, these crucial regulators were absent or functionally altered in the obese mice’s liver networks. Intuitively, such loss might destabilize network structure; however, structural analyses showed remarkable preservation of the network’s overall connectivity, suggesting that obesity impairs metabolic function through subtle temporal rather than structural mechanisms.
To probe these temporal effects, the research team employed comprehensive time-series analyses capturing fluctuations in metabolite concentrations, enzyme activities, and gene expression profiles during starvation. This high-resolution temporal data unveiled a robust temporal hierarchy in healthy livers, where hub molecules swiftly adjusted activity to modulate downstream pathways in an orderly manner. By contrast, in obese mice, this temporal coordination was disorganized, leading to delayed or incomplete metabolic responses despite the intact network architecture.
These findings challenge the conventional paradigm that associates obesity-induced metabolic dysfunction primarily with structural damage to cellular networks. Instead, they highlight the critical importance of temporal dynamics—how effectively and rapidly metabolic responses occur over time. The liver’s temporal vulnerability during starvation in obesity may contribute to impaired energy homeostasis, making it harder for obese organisms to adapt to nutrient deprivation and increasing their susceptibility to metabolic diseases.
The methodology underpinning this research is equally impactful. By combining network structural analysis with meticulous temporal profiling, the team developed a transomic approach that captures both static and dynamic features of metabolic regulation. This framework can be broadly applied to other complex biological systems, integrating data from multiple “omics” layers such as genomics, transcriptomics, and microbiomics. Such integrative analysis promises to unravel the temporal orchestration underlying various physiological and pathological states beyond starvation and obesity.
Kuroda emphasizes the universality of their approach: “Our work provides a global perspective on how metabolic networks adapt—or fail to adapt—to dynamic environmental challenges. Understanding temporal vulnerabilities opens new possibilities for therapeutic interventions targeting timing and coordination rather than solely network components.” Future research may extend these insights to study metabolic responses during food intake cycles, circadian rhythms, and chronic disease progression, potentially transforming how metabolic diseases are diagnosed and treated.
Moreover, this study underscores the limitations of static views in biology. Traditional network analyses often treat molecular interactions as fixed, omitting the crucial influence of timing on biological function. The discovery of temporal vulnerabilities explains why obesity can disrupt metabolism without apparent structural damage, resolving a longstanding paradox in metabolic research. In effect, healthy metabolic regulation emerges not just from network integrity but from precise temporal control over molecular cascades.
The implications of this work extend beyond metabolism. Many complex biological systems—from immune responses to neural networks—rely on tightly coordinated temporal activity. The tools and concepts developed here pave the way for future studies exploring temporal coordination as a fundamental principle of biology and disease. By embracing the fourth dimension—time—scientists can gain a deeper, more actionable understanding of life’s complexity.
In conclusion, the University of Tokyo team’s discovery of temporal dysfunction in the seemingly robust metabolic networks of obese livers revolutionizes our comprehension of starvation response. Their integrated transomic and temporal analytic framework offers an innovative vantage point for studying metabolic adaptation and dysfunction. As the field progresses, this temporal perspective promises to enrich our strategies for addressing obesity and related metabolic disorders, ultimately improving health outcomes through precise temporal targeting of molecular pathways.
Subject of Research: Animals
Article Title: Structural robustness and temporal vulnerability of the starvation-responsive metabolic network in healthy and obese mouse liver
News Publication Date: 22-Apr-2025
Web References: 10.1126/scisignal.ads2547
Image Credits: Morita et al 2025
Keywords
Metabolism, Starvation, Obesity, Liver, Temporal dynamics, Transomic network, Metabolic regulation, Energy homeostasis, Systems biology, Network robustness, Biological timing, Multi-omics analysis
Tags: dynamic temporal analysis of metabolismenergy conservation in obese micegene expression regulation in starvationhomeostasis and obesityliver function during starvationmetabolic adaptation under stressmetabolic networks in micemulti-omics data in researchobesity effects on metabolismstarvation response mechanismstemporal vulnerabilities in metabolic regulationtransomic network in obesity