Bacterial transcription is a production line of copying over genetic instructions from double stranded DNA to produce RNA, which is then used for the subsequent production of proteins that the bacterium needs at that moment. The line supervisors are known as sigma factors, of which there are several varieties that can be largely classified into four groups based on their architecture and genetic sequence.
Credit: LI Jie and FENG Yingang
Bacterial transcription is a production line of copying over genetic instructions from double stranded DNA to produce RNA, which is then used for the subsequent production of proteins that the bacterium needs at that moment. The line supervisors are known as sigma factors, of which there are several varieties that can be largely classified into four groups based on their architecture and genetic sequence.
One factor, however, has been considered “unique.” Called SigI, the factor has a hand in several regulatory responses, but it does not fit into any of the four groups. Now, researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) have discovered that the SigI are more distinct than previously understood — so much so that they require an entirely new classification group.
Their findings, published in Nature Communications on Oct. 13, may help inform the design of gene regulation tools.
In bacterial transcription, sigma factors trigger recognition of a specific promoter — or a region of genetic information right at the start of the DNA sequence that’s needed to produce a specific transcript. The researchers noted that, bafflingly, different SigIs in the same bacterium can bind to different promoters to achieve specific recognition.
Although SigIs have been studied for more than 20 years since their discovery, the molecular mechanism of the specific promoter recognition by SigI during the transcription process had remained unknown. “The existence of multiple SigIs and their role regulating the expression of a super multienzyme complex, called cellulosome, in some bacteria for efficient degradation of plant biomass raised further questions about the mechanism of specificity between different SigIs,” said Prof. FENG Yingang from QIBEBT, co-corresponding author of the study.
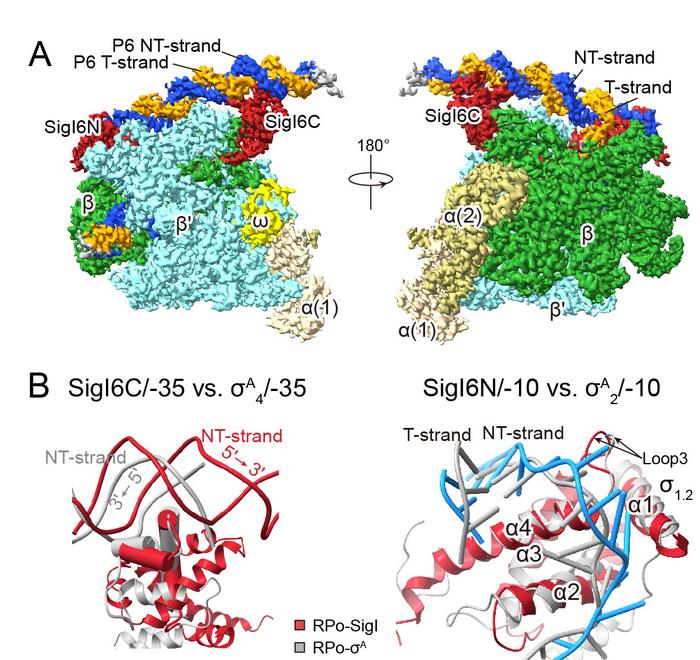
The researchers reconstructed two transcription open complexes formed by two SigI factors, one with SigI1 and one with SigI6, from RNA polymerase purified from the bacteria C. thermocellum with SigI factors purified from E. coli and synthesized promoters. They then used cryo-electron microscopy to determine the complexes’ structures and experimental analysis to test each structure’s function.
From structural and functional analysis, the researchers solved the problems of both SigI classification and the SigI specificity in their functions. They elucidated that SigI factors from a cellulosome-producing bacteria encompass a unique, hitherto unknown recognition mode of bacterial transcriptional promoters and represent a new distinctive class of sigma factors for bacterial transcription.
The structures showed that SigI has two structural features — one of which does not exist in other known sigma factors — that enable a distinct recognition mode for a specific promotor with -35 element.
“SigI promoter recognition of the -35 element differs completely from that of other sigma factors in this family,” FENG said. “The structures also showed that SigIs can recognize another promoter region known as -10 element using different features from the ones used by other sigma factors.”
The team plans to continue investigating the mechanism of SigI factors in the regulation of cellulosomes in C. thermocellum.
“Although we have gained some knowledge of the SigI system in transcriptional regulation of cellulosome, the understanding of cellulosome regulation is still far from complete,” FENG said. “C. thermocellum contains about 80 cellulosomal components, but only a small fraction of them were confirmed to be regulated under the control of the SigI system. Ultimately, our goal is to elucidate the molecular mechanisms governing the regulation of cellulosomes. With this knowledge, we hope to engineer cellulosome-producing bacteria to achieve a better cell factory for applications in bioenergy, synthetic biology and biotechnology.”
Journal
Nature Communications
DOI
10.1038/s41467-023-41796-4
Article Title
Structure of the transcription open complex of distinct σI factors
Article Publication Date
13-Oct-2023




