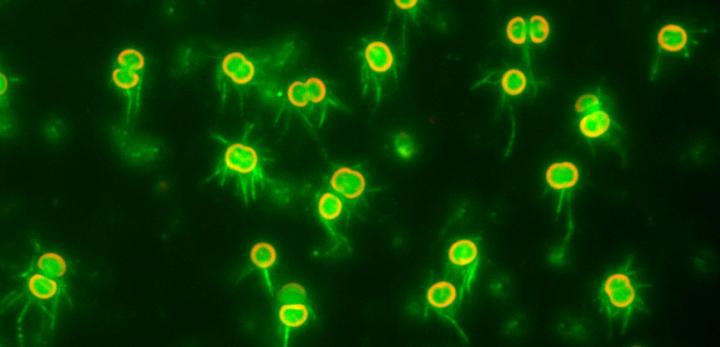
Credit: Nishizaka Lab, Gakushuin University
Type IV pili (T4P) are fascinating supermolecular machines that drive twitching motility, protein secretion, and DNA uptake in prokaryotes. T4P work as grappling hooks that cause bacterial twitching motility by a cycle of extension, surface attachment, and retraction, making the cells move over a surface by pulling themselves along it. The properties of T4P as a motor have previously been scrutinized with biophysics techniques, but the mechanism of regulation of T4P activation dynamics in response to various environmental signals remained unclear, largely because numerous components cooperate to orchestrate the dynamics, and because T4P are very thin (~ 8 nm diameter) and therefore difficult to observe.
In a study published recently in The Proceedings of the National Academy of Science, biophysicists Daisuke Nakane and Takayuki Nishizaka at Gakushuin University in Tokyo overcame these difficulties by developing simple methods to visualize T4P filaments, and used optical microscopy to shed light on the mechanism of regulation of T4P activation dynamics. They directly visualized T4P dynamics at the single cell level in Synechocystis sp. PCC6803, a model cyanobacterium that shows negative phototaxis via twitching motility in response to blue light. T4P can extend from all directions of the body of the essentially spherical Synechocystis cells, and thus the cells can move in any direction along a surface.
Nakane and Nishizaka devised an optical setup that allowed them to simultaneously illuminate specimens on a glass substrate both laterally and vertically. First, they used light microscopy to directly visualize T4P activity as the distribution and intensity of polystyrene beads captured by T4P on the surface of immobilized cells controlled by blue light. The beads were accumulated uniformly on cells illuminated vertically with blue light, but when a cell was partially illuminated with vertical light only on its right half, beads were accumulated only in the illuminated region. When cells were illuminated laterally with blue light, the beads were selectively accumulated on the forward side of the cells (opposite to the light source), indicating elevated T4P activity there. This asymmetric accumulation suggests that T4P filaments located on the forward side of the cell can drive forward motion, since lateral illumination induces negative phototaxis toward this side.
The team then directly visualized and quantified T4P and its dynamics on the cells by labeling cellular T4P with avidin conjugated with FITC and performing microscopic examination of the immobilized cells, and found that accumulation and extension of T4P were also activated at the forward side of cells illuminated laterally with blue light. When they did the same experiment using a mutant that lacks PixD, a blue light-sensing protein, the distribution of T4P in the mutant was more uniform, which suggested that PixD is responsible for the asymmetric distribution of T4P via an unknown mechanism which suppresses the accumulation of T4P on the light-illuminated side.
These observations altogether revealed that the illuminated wild-type cell has an asymmetric distribution of T4P that causes the cell to move away from directional blue light, that this asymmetric distribution is triggered by the local difference of blue-light intensity, and that the T4P extension is activated at the region where the light intensity is higher. These findings appear to be contradictory, because blue light is known to work as a "repellent." However, this could be explained by the previously reported microoptics effect of the cell, namely, when a single cell is exposed to collimated light, the cell condenses the light into the side opposite to the light source because the cell body works as a lens. This strategy seems reasonable because a microorganism with a size on the order of visible wavelengths cannot produce any structure to shield itself from light.
Finally, quantitative cell-tracking analyses of twitching motility and T4P dynamics indicated that 1) T4P filaments were uniformly extended on the cell within 0.2 minutes after blue-light exposure, causing wriggling motion of the cell, 2) T4P then uniformly appeared in all regions of the cell surface, causing random twitching motility of the cell, and 3) within 1 minute after the exposure, the activation of T4P became asymmetric along the light axis in response to the photo-sensing protein PixD, resulting in directional cell motility.
The authors note that their experimental methods could be widely applicable to quantify the dynamic properties of T4P, which may be common in other bacteria such as Neisseria gonorrheae or Pseudomonas aeruginosa, and are perhaps related to the dynamics of archaella (flagella in archaea). The combination of this new experimental system and mutants that lack other light receptors or key proteins in various species will reveal more detailed mechanisms in the near future.
Altogether, these findings highlight how the light-signal processing system in cyanobacteria regulates T4P dynamics to navigate cells in a certain direction. This navigation mechanism is in stark contrast to the mechanism of chemotaxis in bacteria, which operates by changing the interval between random swimming and tumbling.
###
Media Contact
Tomoyuki Matsui
[email protected]
81-359-921-008
@Saku_Gakushuin
http://www.univ.gakushuin.ac.jp/en/
Related Journal Article
http://dx.doi.org/10.1073/pnas.1702395114
############
Story Source: Materials provided by Scienmag