Credit: Tobias Kromann-Hansen
New research provides mechanistic insight into how protein dynamics control the activity of a group of enzymes called serine proteases. As serine proteases play pivotal roles in blood coagulation, the innate immune system and tissue remodeling, the results may be important for the development of new drugs for the treatment of various diseases.
Proteins are usually considered to have a stable three-dimensional structure with a well-defined folding of the peptide chain. However, the peptide chain seems to undergo small or large movements that constantly change the three-dimensional structure of the protein. These constant changes in the three-dimensional structure are termed protein dynamics. Although it is well-established that protein dynamics play a crucial role in the activity level of a group of enzymes called serine proteases, the mechanism behind these observations has remained elusive. As the activity level of serine proteases plays important roles in blood coagulation, the innate immune system and in tissue remodeling, protein dynamics thus become a crucial factor in the regulation of these vital physiological processes.
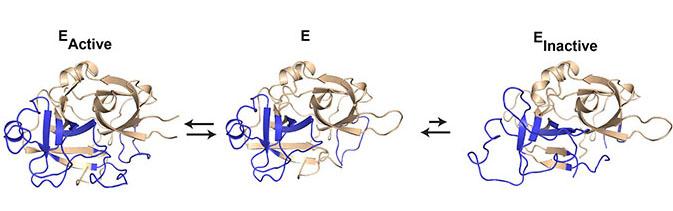
The new research, recently published in the highly recognized journal Nature Scientific Reports, provides a mechanistic insight into the protein dynamics that control the activity level in the serine protease urokinase. The researchers from Aarhus University have solved five different X-ray crystal structures of urokinase.
"Determining the crystal structures of urokinase was far from an easy task," says Postdoc Tobias Kromann-Hansen. "Urokinase is a super dynamic protein that constantly changes its three-dimensional structure. This complicates the formation of crystals," continues Tobias Kromann-Hansen. To solve this problem, the researchers from Aarhus University collaborated with researcher from The Vrije University of Brussels and Leuven University to develop a panel of camelid-derived antibodies that binds specifically to urokinase.
"The camelid antibodies have proven to be a very useful tool in X-ray crystallography as they tend to stabilize dynamic proteins, thus facilitating the formation of crystals," says Tobias Kromann-Hansen. The crystal structures gave the researchers five different snapshots of urokinase and revealed that the urokinase peptide chain undergoes surprisingly large movements. These movements were further mapped using a special technique, HDX-MS (Hydrogen Deuterium Exchange Followed By Mass Spectrometry), in which Professor Elizabeth A. Komives at the University of California, San Diego, is a world-leading expert. Compared with biochemical studies, the X-ray crystal structures and the HDX-MS data provided a detailed description of the molecular mechanisms that underlie the protein dynamics that control the activity level in urokinase.
The results provide a basic understanding of the mechanistic function of serine proteases. But the researchers hope that the results can open up for new possibilities in drug discovery and treatment of various diseases. Tobias Kromann-Hansen explains: "With these results, we have found that urokinase can exist in equilibrium between an active and an inactive state. We now know the form of the inactive state. By developing molecules that specifically recognize and stabilize the inactive state, we may shift the equilibrium towards the inactive state, thereby inhibiting the disease-promoting activity of urokinase in, for example, cancer and arthritis. Our hope is to find similar inactive states in other serine proteases in order to apply this principle in treating other serious diseases that involves a high serine protease activity level."
###
The research was supported by the Carlsberg Foundation through funds for Tobias Kromann-Hansen's postdoc at the University of California San Diego, as well as a grant from the Danish-Chinese Center for Proteases and Cancer, led by former professor Peter A. Andreasen from the Department of Molecular Biology and Genetics at Aarhus University.
Media Contact
Tobias Kromann-Hansen
[email protected]
45-24-45-20-09
@aarhusuni
http://www.au.dk
Original Source
http://mbg.au.dk/en/news-and-events/news-item/artikel/new-knowledge-about-the-dynamics-of-proteins-can-shape-the-future-in-drug-development/ http://dx.doi.org/10.1038/s41598-017-03457-7
############
Story Source: Materials provided by Scienmag