Credit: Steven Offer
Although the development of secondary cancerous growths, called metastasis, is the primary cause of death in most cancers, the cellular changes that drive it are poorly understood. In a new study, published in Genome Biology, researchers at the University of Illinois Urbana-Champaign have developed a new modeling approach to better understand how tumors become aggressive.
“Researchers have identified several cellular pathways that change when a tumor becomes aggressive. However, it is difficult to understand how they affect the tumor,” said Steven Offer, an assistant professor of molecular pharmacology and experimental therapeutics at Mayo Clinic, Minnesota. “We wanted to develop a simple system that can model how cancer cells form an aggressive tumor.”
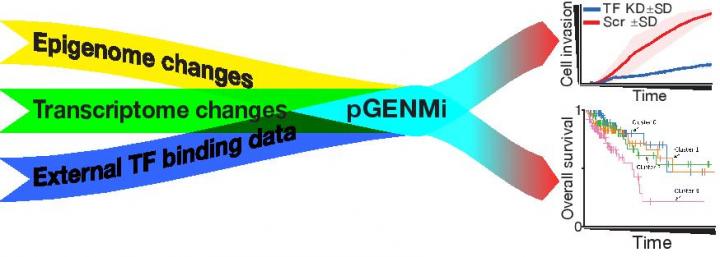
The researchers pooled the data from their own experiments as well as publicly available data to develop the model, which was based on a simpler 2018 model that investigated regulators of cancer drug resistance. In this paper, they specifically focused on transcription factors, which are proteins that control gene expression by binding to the DNA.
“We can easily see how many transcription factors are there in the cancer cell. This model allowed us to see whether the target areas they bind to are available or not,” Offer said. The target areas can be hidden depending on the DNA organization. By studying their availability, the researchers can predict which transcription factors and targets are important.
“The advantage of the model is that it can integrate different types of experimental data, which is not an easy task. It gave us a list of transcription factors, ranked based on their relevance to colorectal cancer aggressiveness,” said Saba Ghaffari, a PhD student in the Sinha lab. The model is so adaptable that it was able to analyze the binding of transcription factors in other types of cells as well.
The researchers also tested the predictions of the model using human cancer cell lines. They looked at the transcription factors that were identified and showed that they were involved in increasing the aggressiveness of the colorectal cancer cells.
“Without the model, it would have been expensive and time consuming for us to analyze the transcription factors in all these different cell lines,” Offer said. “We can now use this data to improve cancer care. The more information we have about these factors, the more disruptions we can create to interfere with the process, and improve treatments.”
The researchers are hoping to improve the model further to make it more sensitive. “Although we binarized the data, the effects of these transcription factors are continuously changing. We also assumed that all the genes work independently of each other; in reality they work together,” Ghaffari said.
“Increasingly, these technologies provide us complementary views of cellular changes during disease progression. Ghaffari’s work provides us with a general-purpose recipe to combine those different views into one meaningful whole, giving us more that any one view can,” said Saurabh Sinha (BSD/CABBI/GNDP/GSP), a professor of computer science. “This is just the beginning. We are looking at it as a blueprint for many more analyses in the future, tackling different biological challenges.”
###
The study “An integrated multi-omics approach to identify regulatory mechanisms in cancer metastatic processes” can be found at 10.1186/s13059-020-02213-x. The work was supported by the NIH, the CompGen Initiative at UIUC, and Mayo Clinic Center for Biomedical Discovery.
Media Contact
Ananya Sen
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




