Reston, VA—A new artificial intelligence tool provides a fully automated, easy-to-use and objective way to detect and evaluate brain tumors, according to new research published in the October issue of The Journal of Nuclear Medicine. Developed for use with amino acid PET scans, the deep learning-based segmentation algorithm can also assess brain tumor patients’ response to treatment with quality comparable to that of an experienced physician, but in a fraction of the time.
Credit: Image created by Robin Gutsche, Institute of Neuroscience and Medicine (INM-4), Research Center Juelich (FZJ), Juelich, Germany.
Reston, VA—A new artificial intelligence tool provides a fully automated, easy-to-use and objective way to detect and evaluate brain tumors, according to new research published in the October issue of The Journal of Nuclear Medicine. Developed for use with amino acid PET scans, the deep learning-based segmentation algorithm can also assess brain tumor patients’ response to treatment with quality comparable to that of an experienced physician, but in a fraction of the time.
PET has become increasingly important in brain tumor diagnostics, complementing structural MRI. Over the past years, several studies have demonstrated the diagnostic value of the metabolic tumor volume for assessing treatment response in brain tumor patients. However, since measuring changes in the metabolic tumor volume of a brain tumor is time-consuming, it is usually not part of the routine clinical assessment.
“The fact that metabolic tumor volume is not routinely assessed in clinical practice suggests that the time and effort required for volumetric amino acid PET segmentation still exceeds clinical benefit,” said Philipp Lohmann, PhD, assistant professor (Habilitation) in Medical Physics, and team leader for Quantitative Image Analysis & AI at the Institute of Neuroscience and Medicine, Research Center in Juelich, Germany. “In response, our team developed a deep learning-based segmentation algorithm for a robust and fully automated volumetric evaluation of amino acid PET data and evaluated its performance for response assessment in patients with gliomas.”
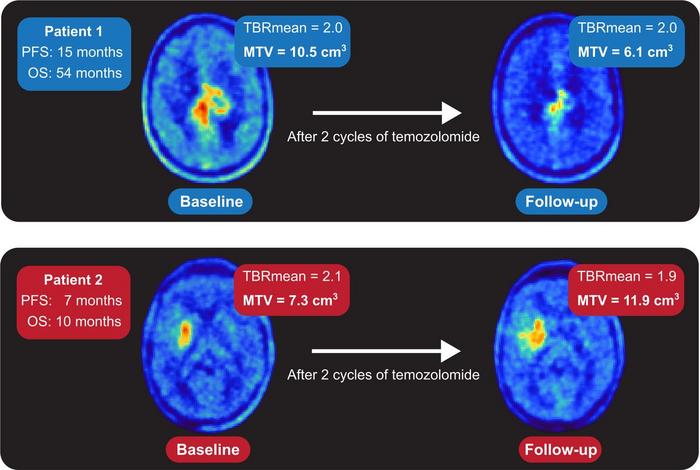
Researchers retrospectively evaluated 699 18F-FET PET scans (at initial diagnosis or during follow-up) from 555 brain tumor patients. The deep learning-based segmentation algorithm was configured on a training and a test dataset, and changes in metabolic tumor volume were measured. Additionally, the algorithm was applied to data from a recently published 18F-FET PET study on response assessment in glioblastoma patients treated with adjuvant temozolomide chemotherapy. The response assessment from the algorithm was then compared to the assessment of an experienced physician, as reported in the study.
In the test dataset, 92 percent of lesions with increased uptake and 85 percent of lesions with isometric or hypometabolic uptake were correctly identified by the algorithm. Change in metabolic tumor volume, as detected by the algorithm, was a significant determinant of disease-free and overall survival, in agreement with the physician’s assessment.
“These findings highlight the value of the deep learning-based segmentation algorithm for improvement and automatization of clinical decision-making based on the volumetric evaluation of amino acid PET,” stated Lohmann. “The segmentation tool developed in our study could be an important platform to further promote amino acid PET and to strengthen its clinical value, which may give brain tumor patients access to important diagnostic information that was previously unavailable or difficult to obtain.”
To facilitate clinical implementation, the segmentation algorithm is freely available and can be executed on a conventional GPU-equipped computer in less than two minutes without preprocessing. “We hope to encourage and support treating physicians in neuro-oncology centers to consider amino acid PET for their patients, even if they have little or no prior experience,” said Lohmann. “Every patient with a brain tumor should have access to amino acid PET.”
This study was made available online in August 2023.
The authors of “Automated Brain Tumor Detection and Segmentation for Treatment Response Assessment Using Amino Acid PET” include Robin Gutsche, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, and RWTH Aachen University, Aachen, Germany; Carsten Lowis and Philipp Lohmann, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany; Karl Ziemons, Medical Engineering and Technomathematics, FH Aachen University of Applied Sciences, Juelich, Germany; Martin Kocher, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, and Department of Stereotaxy and Functional Neurosurgery, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany; Garry Ceccon, Department of Neurology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany; Cláudia Régio Brambilla, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, and JARA-BRAIN-Translational Medicine, Aachen, Germany; Nadim J. Shah, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, JARA-BRAIN-Translational Medicine, Aachen, Germany, and Department of Neurology, University Hospital RWTH Aachen, Aachen, Germany; Karl-Josef Langen, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, Department of Nuclear Medicine, University Hospital RWTH Aachen, Aachen, Germany, and Center for Integrated Oncology, Universities of Aachen, Bonn, Cologne, and Duesseldorf, Germany; Norbert Galldiks, Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH, Juelich, Germany, Department of Neurology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany, and Center for Integrated Oncology, Universities of Aachen, Bonn, Cologne, and Duesseldorf, Germany; and Fabian Isensee, Applied Computer Vision Lab, Helmholtz Imaging, Heidelberg, Germany, and Division of Medical Image Computing, German Cancer Research Center, Heidelberg, Germany.
Visit the JNM website for the latest research, and follow our new Twitter and Facebook pages @JournalofNucMed or follow us on LinkedIn.
###
Please visit the SNMMI Media Center for more information about molecular imaging and precision imaging. To schedule an interview with the researchers, please contact Rebecca Maxey at (703) 652-6772 or [email protected].
About JNM and the Society of Nuclear Medicine and Molecular Imaging
The Journal of Nuclear Medicine (JNM) is the world’s leading nuclear medicine, molecular imaging and theranostics journal, accessed 15 million times each year by practitioners around the globe, providing them with the information they need to advance this rapidly expanding field. Current and past issues of The Journal of Nuclear Medicine can be found online at http://jnm.snmjournals.org.
JNM is published by the Society of Nuclear Medicine and Molecular Imaging (SNMMI), an international scientific and medical organization dedicated to advancing nuclear medicine and molecular imaging—precision medicine that allows diagnosis and treatment to be tailored to individual patients in order to achieve the best possible outcomes. For more information, visit www.snmmi.org.
Journal
Journal of Nuclear Medicine
DOI
10.2967/jnumed.123.265725
Article Title
Automated Brain Tumor Detection and Segmentation for Treatment Response Assessment Using Amino Acid PET
Article Publication Date
1-Oct-2023
COI Statement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; projects 428090865/SPP 2177 [Robin Gutsche, Norbert Galldiks, and Philipp Lohmann] and 491111487). Part of this work was funded by Helmholtz Imaging, a platform of the Helmholtz “Incubator on Information and Data Science.” Norbert Galldiks and Philipp Lohmann received honoraria for lectures from Blue Earth Diagnostics. Norbert Galldiks received honoraria for advisory board participation from Telix Pharmaceuticals.



