New research from UNC Charlotte’s Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER) has found that the two recent and prevalent strains of the virus that cause COVID-19, SARS-CoV-2 variants BA.2.86 and JN.1, are not significantly better than their predecessor Omicron at evading immune responses and causing infections despite having a high number of mutations compared to Omicron.
Credit: UNC Charlotte
New research from UNC Charlotte’s Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER) has found that the two recent and prevalent strains of the virus that cause COVID-19, SARS-CoV-2 variants BA.2.86 and JN.1, are not significantly better than their predecessor Omicron at evading immune responses and causing infections despite having a high number of mutations compared to Omicron.
When first identified, the Omicron variant, BA.2.86, and its close relative, JN.1, raised significant public health concerns. These concerns were tied to the fact that the original Omicron variant was highly mutated, resulting in both immune evasion and breakthrough infections, as well as being more infectious and highly-mutated compared to earlier variants.
There was some speculation that large numbers of new mutations in BA.2.86 and JN.1 conferred a greater ability of these variants to evade the human immune system and be more transmissible. Extensive computational analyses conducted by a team of UNC Charlotte scholars and students determined that these variants only had small, statistically insignificant changes in immune evasion and host-cell binding capacity compared to earlier variants, including Omicron.
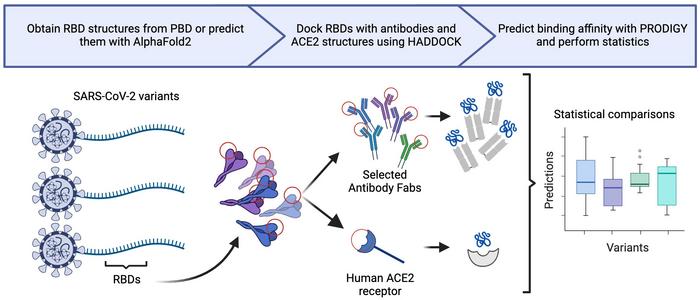
To assess the immune evasion of BA.2.86 and JN.1, the UNC Charlotte research team performed in silico analyses on the Receptor Binding Domain (RBD; the region of the viral genome against which mRNA vaccines are designed) of SARS-CoV-2, comparing the two newer variants to previous variants to calculate the relative binding affinity of neutralizing antibodies (from vaccinated patients, infected patients, and therapeutic sources) to the RBD. In addition to antibody analyses, researchers calculated the relative binding affinity of BA.2.86 and JN.1 to Angiotensin Converting Enzyme-2 (ACE2; the viral receptor on human cells) in comparison to previous variants.
The team found minor changes in binding affinity for neutralizing antibodies and ACE2 for BA.2.86 and JN.1 in comparison to previous SARS-CoV-2 variants. However, those changes were not statistically significant. Therefore, they concluded that BA.2.86 and JN.1 have no significant increase in immune evasion or transmissibility to previous variants.
The viral RBD is critical to bind to the ACE2 receptor of the human cell, making it improbable that the RBD will drastically change to evade existing neutralizing antibodies induced by vaccine or previous infection without significantly decreasing the RBD’s affinity to ACE2.
Many of the RBD residues critical to binding to ACE2 are also targets of antibodies. Neutralizing antibodies that target the RBD have become increasingly specific and efficacious. As a result of this biochemical stalemate, proteins in SARS-CoV-2 outside of the RBD have accumulated mutations as new variants have arisen.
For example, the JN.1 variant has three mutations outside of the RBD relative to its recent predecessor BA.2.86. These mutations remain to be studied but early data indicate that they increase viral replication and down regulate the host cell’s immune system. In the past, some of these mutations within these proteins have led to enhanced immune evasion and increased viral replication. Thus, these mutations may have allowed JN.1 to outcompete BA.2.86 in its prevalence among SARS-CoV-2 variants.
“We believe RBD evolution is becoming asymptotic,” said Shirish Yasa, a research assistant in CIPHER. “Evolutionary pressure has now turned towards those genes outside of the RBD. As a result, future research will focus on the genes outside of the RBD.”
“We are entering a whole new era of molecular epidemiology by demonstrating the need to add a functional approach to viral nucleotide sequencing efforts. Although Omicron was highly mutated, immune evasive, and created a severe public health burden, our results for JN.1 and BA.2.86 are in contrast. Taken together, our results demonstrate that counting the mutations of the RBD alone does not indicate immune evasion and possible new burdens on public health,” said Daniel Janies, the co-director of CIPHER and the Carol Grotnes Belk Distinguished Professor of Bioinformatics and Genomics in the College of Computing and Informatics.
The authors include:
- Shirish Yasa
- Sayal Guirales-Medrano
- Denis Jacob Machado
- Colby T. Ford
- Daniel Janies
Yasa, S., Guirales-Medrano, S., Jacob Machado, D, Ford, C.T., Janies, D. 2024. Predicting Antibody and ACE2 Affinity for SARS-CoV-2 BA.2.86 and JN.1 with In Silico Protein Modeling and Docking. Frontiers in Virology, 4, https://www.frontiersin.org/journals/virology/articles/10.3389/fviro.2024.1419276
DOI=10.3389/fviro.2024.1419276
ISSN=2673-818X
Journal
Frontiers in Virology
DOI
10.3389/fviro.2024.1419276
Method of Research
Computational simulation/modeling
Article Title
Predicting Antibody and ACE2 Affinity for SARS-CoV-2 BA.2.86 and JN.1 with In Silico Protein Modeling and Docking
Article Publication Date
19-Jul-2024




