Credit: Jacob Rosenstein et. al.
PROVIDENCE, R.I. [Brown University] — DNA molecules are well known as carriers of huge amounts of biological information, and there is growing interest in using DNA in engineered data storage devices that can hold vastly more data than our current hard drives. But new research shows that DNA isn’t the only game in town when it comes to molecular data storage.
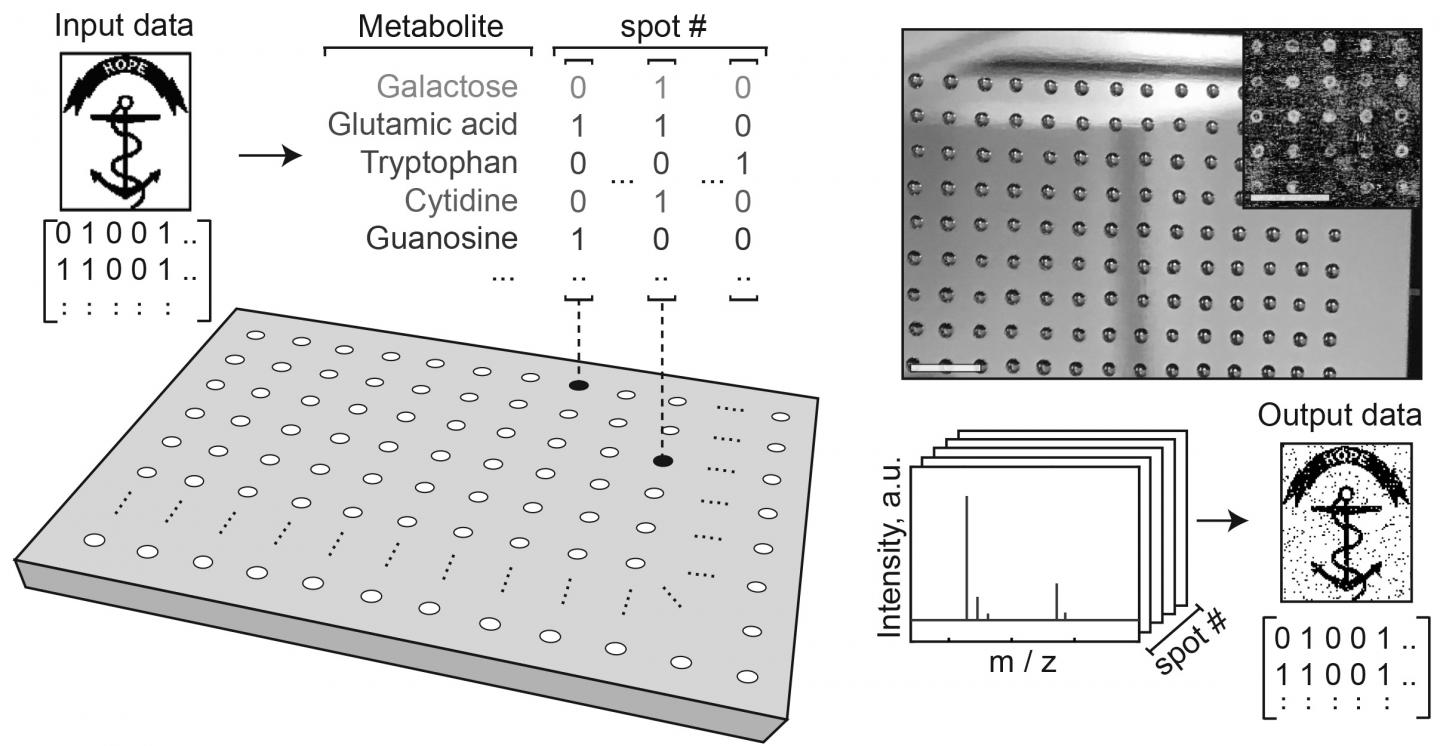
A study led by Brown University researchers shows that it’s possible to store and retrieve data stored in artificial metabolomes — arrays of liquid mixtures containing sugars, amino acids and other types of small molecules. For a paper published in the journal PLOS ONE, the researchers showed that they could encode kilobyte-scale image files into metabolite solutions and read the information back out again.
“This is a proof-of-concept that we hope makes people think about using wider ranges of molecules to store information,” said Jacob Rosenstein, a professor in Brown’s School of Engineering and senior author of the study. “In some situations, small molecules like the ones we used here can have even greater information density than DNA.”
Another potential advantage, Rosenstein says, stems from the fact that many metabolites can react with each other to form new compounds. That creates the potential for molecular systems that not only store data, but also manipulate it — performing computations within metabolite mixtures.
The idea behind molecular computing grows out of an increasing need for more data storage capacity. By 2040, the world will have produced as much as 3 septillion (that’s 3 followed by 24 zeros) bits of data by some estimates. Storing, searching and processing all of that data is a daunting challenge, and there simply may not be enough chip-grade silicon on Earth to do this with traditional semiconductor chips. Funded by a contract with the Defense Advanced Research Projects Administration (DARPA), a group of engineers and chemists at Brown has been working on a variety of techniques for using small molecules to create new information systems.
For this new study, the group wanted to see if artificial metabolomes could be a data-storage option. In biology, a metabolome is the full array of molecules an organism uses to regulate its metabolism.
“It’s not hard to recognize that cells and organisms use small molecules to transmit information, but it can be harder to generalize and quantify,” said Eamonn Kennedy, a postdoctoral associate at Brown and first author of the study. “We wanted to demonstrate how a metabolome can encode precise digital information.”
The researchers assembled their own artificial metabolomes — small liquid mixtures with different combinations of molecules. The presence or absence of a particular metabolite in a mixture encodes one bit of digital data, a zero or a one. The number of molecule types in the artificial metabolome determines the number of bits each mixture can hold. For this study, the researchers created libraries of six and 12 metabolites, meaning each mixture could encode either six or 12 bits. Thousands of mixtures are then arrayed on small metal plates in the form of nanoliter-sized droplets. The contents and arrangement of the droplets, precisely placed by a liquid-handling robot, encodes the desired data.
The plates are then dried, leaving tiny spots of metabolite molecules, each holding digital information. The data can then be read out using a mass spectrometer, which can identify the metabolites present at each spot on the plate and decode the data.
The researchers used the technique to successfully encode and retrieve a variety of image files of sizes up to 2 kilobytes. That’s not big compared to the capacity of modern storage systems, but it’s a solid proof-of-concept, the researchers say. And there’s plenty of potential for scaling up. The number of bits in a mixture increases with the number of metabolites in an artificial metabolome, and there are thousands of known metabolites available for use.
There are some limitations, the researchers point out. For example, many metabolites chemically interact with each other when placed in the same solution, and that could result in errors or loss of data. But that’s a bug that could ultimately become a feature. It may be possible to harness those reactions to manipulate data — performing in-solution computations.
“Using molecules for computation is a tremendous opportunity, and we are only starting to figure out how to take advantage of it,” said Brenda Rubenstein, a Brown assistant professor of chemistry and co-author of the study.
“Research like this challenges what people see as being possible in molecular data systems,” Rosenstein said. “DNA is not the only molecule that can be used to store and process information. It’s exciting to recognize that there are other possibilities out there with great potential.”
###
Other authors on the paper are Christopher Arcadia, Joseph Geiser, Peter Weber and Christopher Rose. The research was supported by DARPA (W911NF-18-2-0031).
Media Contact
Kevin Stacey
[email protected]
Related Journal Article
http://dx.