Credit: JING Gongchao
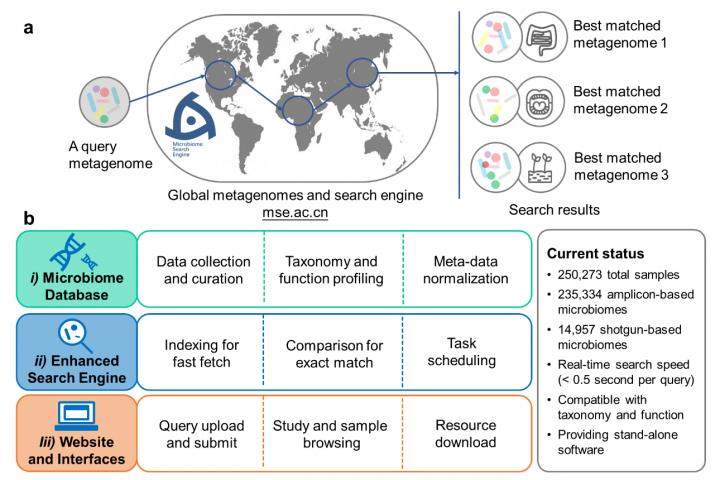
Metagenomics – the study of genetic material from an environmental sample – is growing as species evolve or are discovered across the globe. To correlate the newly developed microbiomes with existing data sets, a team of researchers based in China has developed the Microbiome Search Engine 2 (MSE 2). It was published on Jan. 19 in mSystems, a journal of the American Society for Microbiology.
“Here, we introduce MSE 2, a microbiome database platform for searching query microbiomes in the global metagenome data space based on taxonomic or functional similarity of the whole microbiome,” said co-first author JING Gongchao, Single-Cell Center, CAS Key Laboratory of Biofuels, Qingdao Institute of BioEnergy and Bioprocess Technology (QIBEBT), Chinese Academy of Sciences (CAS).
The previous version of the search engine limited queries to taxonomical similarities, meaning the functional genes had to match. There was not a way for researchers to compare different samples that performed the same function in various microbiomes. The ability to study how the same function may have evolved in different microbes could offer guidance in identifying and treating diseases, according to JING.
“A search-based strategy is useful for large-scale mining of microbiome datasets, providing a bird’s eye view of the microbiome data space and disease diagnosis via microbiome big data,” JING said.
“The new ability to search the microbiome space via functional similarity greatly expands the scope of search-based mining of the microbiome big data,” JING added. “By adding a function-based dimension for these and related applications, MSE 2 should accelerate large-scale mining of the ever-expanding metagenome data space.”
MSE 2 includes an extended database with meta data from 819 studies, updated data compatibility to better incorporate newly available data sets and a user-friendly interface. A single query takes less than half of a second to search against the entire database of more than 260,000 samples.
“Over the past decade we have been passionately collecting published microbiome data – they record the kinds of microbe species and the types of microbial communities that have ever lived on our planet. By collecting and curating them in a minable database, MSE 2 allows these invisible but pivotal creatures on Earth to be “remembered” by future generations, and those scientists who first discovered them to be recognized”, said co-first author LIU Lu, also from Single-Cell Center.
###
MSE 2 was developed by a team led by SU Xiaoquan from College of Computer Science and Technology, Qingdao University, and XU Jian from Single-Cell Center, QIBEBT, CAS. The Natural Science Foundation of China and the Shandong Natural Science Foundation supported this work.
Media Contact
CHENG Jing
[email protected]
Original Source
http://english.
Related Journal Article
http://dx.




