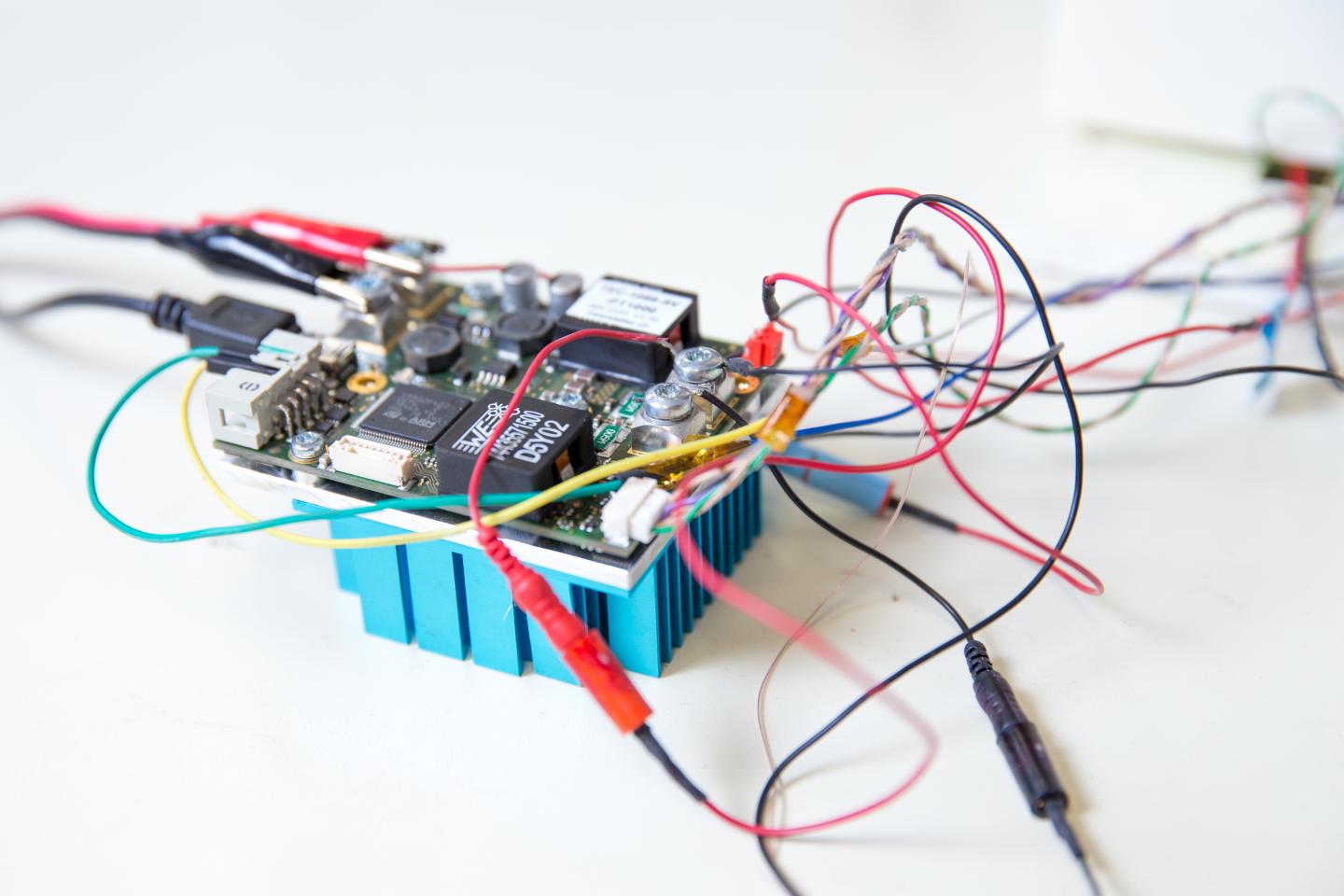
Credit: Jacobs School of Engineering/UC San Diego
Engineers at the University of California San Diego have developed a desktop diagnosis tool that detects the presence of harmful bacteria in a blood sample in a matter of hours instead of days. The breakthrough was made possible by a combination of proprietary chemistry, innovative electrical engineering and high-end imaging and analysis techniques powered by machine learning. The team details their work in the Feb. 8 issue of Scientific Reports.
To identify low levels of harmful bacteria among a large number of human blood cells, researchers for the first time melted bacterial DNA in 20,000 extremely small simultaneous reactions. Each reaction contained only 20 picoliters–a scale that is hard to picture: one drop of rain contains hundreds of thousands of picoliters.
Each type of DNA has a specific signature as it comes apart during melting. As the melting process is imaged and analyzed, researchers can use machine learning to determine which types of DNA appear in blood samples. During experiments, the system accurately identified, 99 percent of the time, DNA sequences from bacteria causing food-borne illnesses and pneumonia–in less than four hours.
"Analyzing this many reactions at the same time at this small a scale had never been attempted before," said Stephanie Fraley, a professor of bioengineering at the Jacobs School of Engineering at UC San Diego and the paper's lead author. "Most molecular tests look at DNA on a much larger scale and look for just one type of bacteria at a time. We analyze all the bacteria in a sample. This is a much more holistic approach."
Current methods used to detect and identify bacteria rely on cultures, which can take days. That is too long to provide physicians with an effective and timely diagnosis tool–as anyone who has been prescribed antibiotics while waiting for test results knows.
"We are driven by clinical needs," Fraley said.
She brought together a team of bioengineers, clinicians, electrical engineers and computer scientists to develop a faster diagnosis system.
How it works
It all starts with one milliliter of blood, which researchers inoculated with Listeria monocytogenes, a food-borne bacterium that causes about 260 deaths a year in the United States, and Streptococcus pneumoniae, which causes everything from sinus infections, to pneumonia, to meningitis.
Researchers isolated all DNA from the blood sample. The DNA was then placed on a digital chip that allowed each piece to independently multiply in its own small reaction. For the process to work at such small scales–each well containing DNA in the chip was only 20 picoliters in volume–researchers used a proprietary mix of chemicals subject to a provisional patent.
The chip with the amplified DNA was placed in an innovative high-throughput microscope that Fraley and her team designed. The DNA was then heated in increments of 0.2 degrees Celsius, causing it to melt at temperatures between 50 to 90 degrees Celsius -about 120 to 190 degrees Fahrenheit.
As the DNA double-helix melts, the bonds holding together the DNA strands break. Depending on the DNA's sequence, the bonds have different strengths and that changes the way the strands unwind from each other. This creates a unique sequence-dependent fingerprint, which researchers can detect using a special dye. The dye causes the unwinding process to give off fluorescent light, creating what researchers call a melting curve–a unique signature for each type of bacteria.
When engineers imaged the melting process with the high-throughput microscope, they were able to capture the bacteria's melting curves. They then analyzed the curves with a machine learning algorithm they developed.
In previous work, the algorithm was trained on 37 different types of bacteria undergoing different reactions in different conditions. The researchers showed that it was able to identify bacteria strains with 99 percent accuracy. By contrast, the error rate for traditional methods can be up to 22.6 percent.
Next steps
Next steps include shrinking the size of the system so that it can be more easily deployed in clinics and physicians' offices. Researchers also want to add to the system the capability to detect fungal and viral pathogens, as well as genes for antibiotic resistance. They also want to further validate their results on patient samples.
Fraley hopes the system will be available to physicians in the next five years.
"This has the potential to reach people near or at the point of care," she said. "With further improvements, it could also be deployed in low-resources settings. It's a simple and innovative approach."
In addition to Fraley, authors are: Daniel Velez Ortiz, Hannah Mack, Julietta Jupe, Sinead Hawker and Yang Zhang, graduate students in the Department of Bioengineering at UC San Diego; Ninad Kulkarni and Behnam Hedayatnia, from the Department of Electrical and Computer Engineering at UC San Diego; and Dr. Shelley Lawrence, an assistant professor of pediatrics at the UC San Diego School of Medicine and Rady Children's Hospital of San Diego.
###
The research was supported by a UC San Diego Clinical and Translational Research Institute Pilot Grant, the Burroughs Wellcome Fund Career Award at the Scientific Interface and the UC San Diego Frontiers of Innovation Scholars Program.
LINK TO THE PAPER TO COME
Media Contact
IOANA PATRINGENARU
[email protected]
858-822-0899
@UCSanDiego
http://www.ucsd.edu