Credit: NINS/IMS
Biomolecular motors in cells generate unidirectional motion, consuming chemical energy gained by, for example, hydrolysis of ATP. Elucidation of the operation principle of such molecular motors, which are nature-made nanomachines composed of proteins, has attracted much attention. Single-molecule imaging, which can directly capture the motion of molecular motors, is a promising technique toward understanding the operation principle of molecular motors. However, it is still unclear how the consumption of chemical energy, i.e., change in chemical states of such motor proteins, gives rise to the unidirectional motion of the entire motors. Researchers at Institute for Molecular Science and Shizuoka University have found the change of the shapes of free energy profiles along the motion of a molecular motor triggered by chemical state changes of the motor.
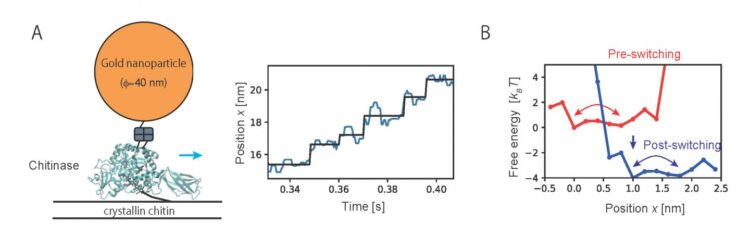
The researchers first tried to establish a computational model to describe the motion of the molecular motors. The motion of a motor can be regarded as diffusive motion on free energy profiles that switch according to the chemical states of molecules which consist of the motor. More specifically, as shown in Fig. 1A, the motor first moves on the free energy surface of chemical state 1 (red) of the motor molecules, and then moves on the free energy surface of chemical state 2 (blue). However, this chemical state switching is not usually observed in single-molecule imaging. The researchers treated the transition between the chemical states using a hidden Markov model in which the chemical states are regarded as “hidden” states (Fig. 1B).
Using this hidden Markov model, it is possible to calculate “likelihood”, which evaluates the probability to show how well the model explains the trajectory of the actual single-molecule motion. It is also possible to incorporate knowledge of the free energy profiles as prior probabilities. The researchers have developed a method for estimating chemical-state-dependent free energy profiles, diffusion coefficients on each profile, and rate constants of transitions between these states within the Bayesian inference framework by Monte Carlo sampling using posterior probabilities expressed as a product of the likelihood and the prior probabilities.
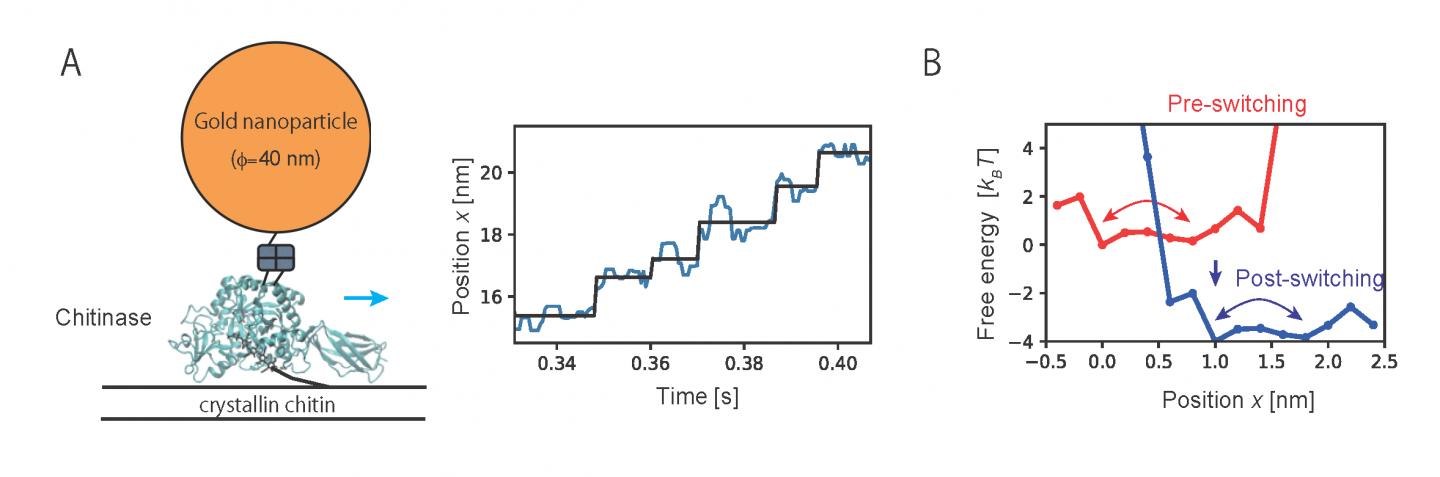
Then, the method developed in the present study was applied to analyze the motion of chitinase, a linear molecular motor, observed by single-molecule imaging. Analyzing trajectory data of unidirectional motion of chitinase with degrading a chitin chain revealed the characteristic free energy profiles which govern the motion (Fig. 2). Results of the analysis showed that a chitinase gets on a rail of chitin chain over a relatively low free energy barrier by Brownian motion. Then, the unidirectional motion is achieved by switching chemical states through the hydrolysis reaction of the chitin chain and dissociation of the reaction products. The present study provides a physical basis for the “burnt-bridge” Brownian ratchet mechanism* that the researchers have previously reported.
“We will apply our method developed in this study to various molecular motors and hope to clarify the similarities and differences in the mechanisms of the molecular motors. We believe that new findings will be obtained by our method in the future and give us a clue to the general operation principles of molecular motors. Studies using our method will pave the way for designing new artificial molecular motors,” said Okazaki.
* “Burnt-bridge” Brownian ratchet mechanism:
A mechanism to explain how to achieve unidirectional motion of a molecular motor using nondirectional Brownian motion due to thermal fluctuations as driving force. In this mechanism the backward motion is prevented by removing the rail behind the molecular motor. (Akihiko Nakamura et al. in Nature Communications (2018) DOI: 10.1038/s41467-018-06362-3. See also EurekAlert! Research News URL: https:/
###
Media Contact
Kei-ichi Okazaki
[email protected]
Related Journal Article
http://dx.