In a groundbreaking study recently published in Science Advances, researchers from the University of Cambridge’s Sainsbury Laboratory have uncovered intricate molecular mechanisms that explain how plants regulate their growth and development through the nuanced functions of SCAR/WAVE proteins. This discovery highlights not only the complexity of cellular control in plants but also reveals how closely related proteins may diverge drastically in their biological roles through subtle sequence variations, which could revolutionize our understanding of plant cell biology and open new pathways for agricultural innovation.
Central to plant cell development is the cytoskeleton, a dynamic internal scaffold primarily composed of actin filaments that shape cellular structures and enable growth processes. The SCAR/WAVE protein complex plays a pivotal role in this architecture by orchestrating the activation of the ARP2/3 complex. This activation initiates the nucleation and branching of actin filaments, facilitating cellular morphogenesis critical for many plant functions. Although SCAR/WAVE genes are widely conserved across plant species, they frequently appear as small gene families encoding similar yet functionally distinct paralogs, posing longstanding questions about their specific biological roles.
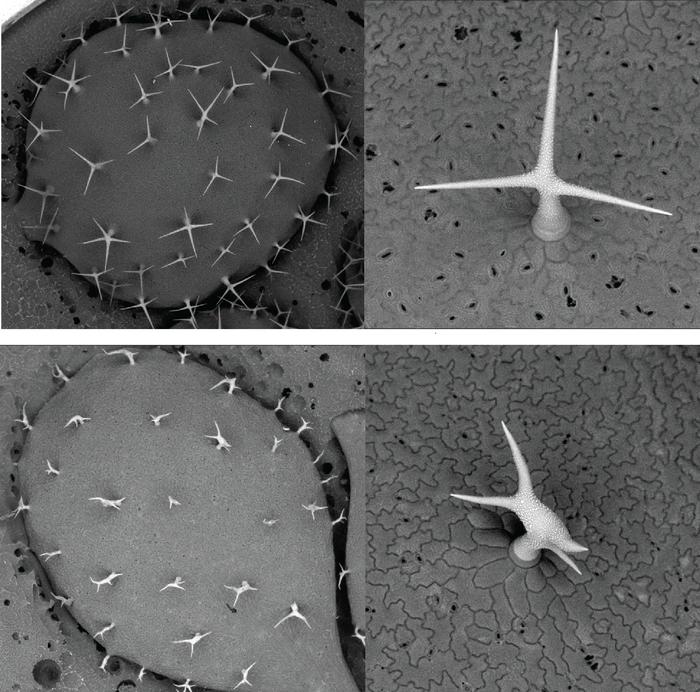
The Cambridge research team directed by Dr. Sebastian Schornack focused their investigations on two related SCAR proteins, MtAPI and MtHAPI1, in the widely studied model legume Medicago truncatula. Unlike previous assumptions that such paralogs might be functionally redundant, their experiments revealed a striking functional divergence. MtAPI was found indispensable for root hair development—a process vital for nutrient and water absorption from soil—while MtHAPI1 was unable to substitute for this purpose. Conversely, when these proteins were expressed in Arabidopsis thaliana, a classic model organism with well-characterized trichome development pathways, MtHAPI1 could compensate for defects in leaf hair formation caused by a defective Arabidopsis SCAR protein, whereas MtAPI could not. These findings provide irrefutable evidence that these homologous proteins have evolved distinct specialized functions within the plant kingdom.
Delving deeper into the molecular basis for this divergence, the researchers uncovered a remarkably influential 42-amino acid sequence embedded within an intrinsically disordered region (IDR) of the MtAPI protein. Intrinsically disordered regions are protein segments that lack a defined three-dimensional structure but are known to mediate versatile interactions and regulatory functions. This particular sequence was demonstrated to substantially reduce the stability of MtAPI by promoting its degradation within plant cells. Intriguingly, this destabilizing effect was transferable and exerted a similar influence when fused onto other, unrelated proteins, even across different species, indicating a broad, evolutionarily conserved regulatory mechanism for controlling protein abundance in plants.
The role of SCAR/WAVE complexes in actin cytoskeleton dynamics is foundational to cellular morphogenesis, and this study’s revelation that functional specificity arises from IDRs fundamentally changes how researchers perceive protein regulation in plants. The subtle tuning of protein levels through these disordered regions suggests an elegant mechanism by which plants can fine-tune developmental processes like root hair elongation and trichome morphology, optimizing adaptation to diverse environmental conditions. This regulatory versatility might allow plants to balance growth demands with resource availability in fluctuating environments.
Dr. Sabine Brumm, lead author of the study, emphasized the significance of these findings by highlighting the importance of intrinsically disordered regions: “Our work shines a light on an often-overlooked facet of protein function. These flexible regions are crucial for defining the functionalities of SCAR proteins, enabling plants to use closely related molecules in unique ways depending on developmental needs.” By reframing the understanding of protein function away from static structural motifs to dynamic sequence elements, this research paves the way for innovative approaches to manipulating plant growth at the molecular level.
This study also elucidates the complex interplay between paralogous proteins that arise from gene duplication events—a source of genetic diversity in plants. The discovery that such paralogs can evolve key sequence variations in IDRs to fine-tune their stability and function exemplifies evolutionary ingenuity. This finding expands the conceptual framework for investigating gene family evolution, particularly in relation to the regulation of protein abundance and functional diversification.
From an applied perspective, understanding how specific SCAR/WAVE proteins influence root hair and trichome development opens promising avenues for agricultural biotechnology. For instance, enhancing root hair growth through stable expression of particular SCAR variants could improve nutrient uptake efficiency, reducing fertilizer dependency and increasing crop yield sustainability. Likewise, modifying trichome development could strengthen physical barriers against pests and environmental stresses, contributing to natural plant defense strategies.
The implications of this research extend beyond developmental biology, intersecting with plant-microbe interactions and environmental adaptability. Since root hairs directly interact with soil microbes, manipulating SCAR protein dynamics might influence symbiotic relationships or pathogen resistance. Accordingly, this study not only enriches fundamental scientific knowledge but also lays groundwork for translational research aimed at meeting global food security challenges under climate change pressures.
As the field moves forward, future studies will undoubtedly focus on further characterizing the role of intrinsically disordered regions in other protein complexes and identifying additional sequence motifs governing protein stability. Moreover, exploring how such regulatory elements respond to environmental cues could unravel new layers of cellular control, enabling fine-tuned and dynamic modifications of plant structure and function in real time.
In conclusion, the work from the University of Cambridge offers a remarkable glimpse into the molecular sophistication of plant developmental regulation. By revealing how intrinsic sequence disorder mediates the functional diversification of SCAR/WAVE proteins, this study provides a conceptual breakthrough with far-reaching implications for biology, agriculture, and beyond.
Subject of Research: Cells
Article Title: Functional divergence of plant SCAR/WAVE proteins is determined by intrinsically disordered regions
News Publication Date: 21-May-2025
Web References:
https://doi.org/10.1126/sciadv.adt6107
References:
Sabine Brumm, Aleksandr Gavrin, Matthew MacLeod, Guillaume Chesneau, Annika Usländer and Sebastian Schornack (2025) Functional divergence of plant SCAR/WAVE proteins is determined by intrinsically disordered regions, Science Advances. DOI: 10.1126/sciadv.adt6107
Image Credits: Sabine Brumm
Keywords: Developmental biology, Cell development, Developmental genetics, Plant development
Tags: actin filament dynamics in plantsagricultural innovation through plant biologyCambridge University plant researchcellular morphogenesis in plantscytoskeleton in plant cellsfunctional diversity of SCAR proteinsgene family evolution in plantsmechanisms of plant growth regulationmolecular biology of plant developmentplant cell architecture and structureplant cell growth mechanismsSCAR/WAVE protein functions