A Kanazawa University researcher finds a way to accelerate a fundamental problem in computing known as non-rigid point set registration
Credit: Kanazawa University
Kanazawa, Japan – Non-rigid point set registration is the process of finding a spatial transformation that aligns two shapes represented as a set of data points. It has extensive applications in areas such as autonomous driving, medical imaging, and robotic manipulation. Now, a method has been developed to speed up this procedure.
In a study published in IEEE Transactions on Pattern Analysis and Machine Intelligence, a researcher from Kanazawa University has demonstrated a technique that reduces the computing time for non-rigid point set registration relative to other approaches.
Previous methods to accelerate this process have been computationally efficient only for shapes described by small point sets (containing fewer than 100,000 points). Consequently, the use of such approaches in applications has been limited. This latest research aimed to address this drawback.
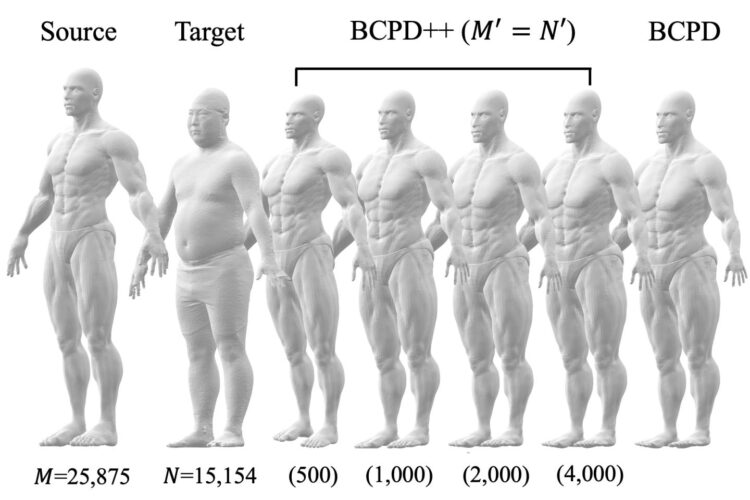
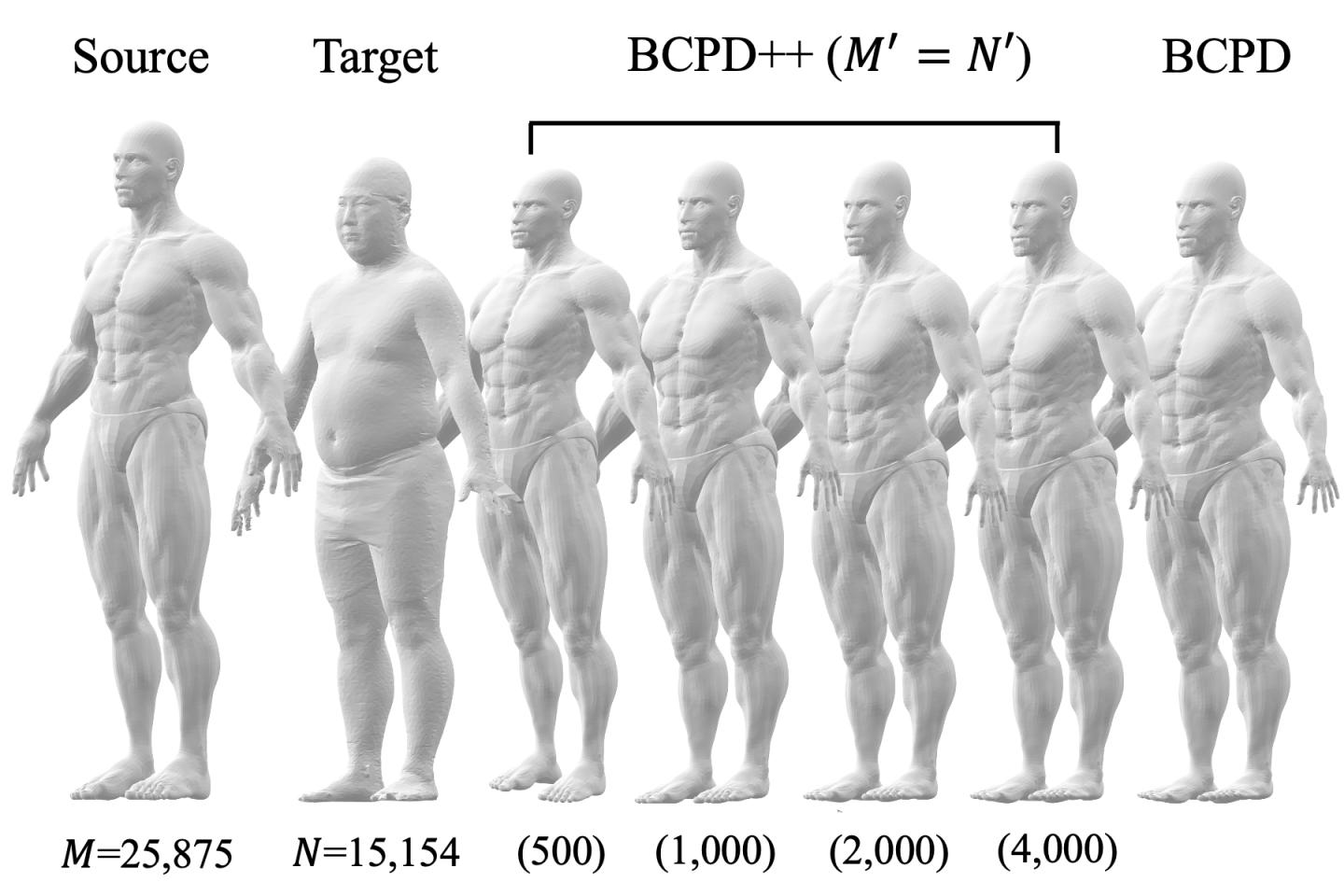
The proposed method consists of three steps. First, the number of points in each point set is reduced through a procedure called downsampling. Second, non-rigid point set registration is applied to the downsampled point sets. And third, shape deformation vectors–mathematical objects that define the desired spatial transformation–are estimated for the points removed during downsampling.
“The downsampled point sets are registered by applying an algorithm known as Bayesian coherent point drift,” explains author Osamu Hirose. “The deformation vectors corresponding to the removed points are then interpolated using a technique called Gaussian process regression.”
The researcher carried out a series of experiments to compare the registration performance of their method with that of other approaches. They considered a wide variety of shapes, some described by small point sets and others by large point sets (containing from 100,000 to more than 10 million points). These shapes included, for example, that of a dragon, a monkey, and a human.
The results demonstrate that the proposed technique is efficient even for point sets with more than 10 million points, shown in Fig. 2. They also show that the computing times of this method are noticeably shorter than those of a state-of-the-art approach for point sets with more than a million points.
“Although the new technique provides accelerated registration, it is relatively sensitive to artificial disturbances in small data sets,” says Hirose. “Such sensitivity indicates that the approach is best suited for large point sets, as opposed to small, noisy ones.”
Given that non-rigid point set registration has a wide range of applications, the method established in this study could have far-reaching implications. The source code of the proposed method is distributed by the author at https:/
###
Media Contact
Tomoya Sato
[email protected]
Original Source
https:/
Related Journal Article
http://dx.