In the rapidly evolving field of graph neural networks, a significant breakthrough has emerged with the introduction of Dynamic Disentangled Graph Convolutional Networks, affectionately dubbed D2-GCN. This innovative approach seeks to enhance node classification tasks by cleverly addressing limitations in traditional Graph Convolutional Networks (GCNs). Classic GCNs typically aggregate features from neighboring nodes in a holistic manner, often neglecting the diverse influences that different neighbors may exert on a node’s representation. As a result, this holistic perspective may lead to a loss of crucial nuances in understanding individual node characteristics.
The research team led by Chuliang Weng has made strides in rectifying these issues by proposing D2-GCN, a powerful model that tailors the number of disentangled feature channels for each node during the training process. This hyper-adaptive approach aims to produce more precise and versatile representations of nodes across different datasets. With D2-GCN, nodes are not merely viewed as static aggregates, but rather as dynamic entities whose representations evolve in response to their surrounding graph structures.
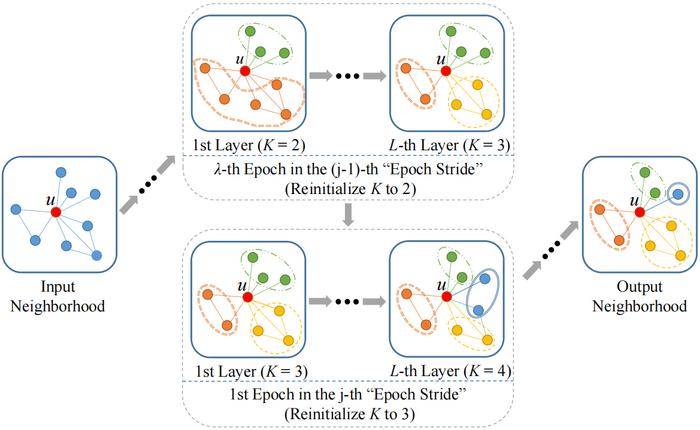
At the heart of the D2-GCN construction lies a sophisticated two-level disentangling mechanism. This mechanism intricately combines epoch-level and layer-level disentanglement during the training sessions, enhancing the model’s capacity to capture subtle changes in node representations. This is particularly valuable when dealing with graphs exhibiting varying topological complexities, where node relationships may fluctuate dramatically across epochs or layers.
Further contributing to the model’s robustness is the expectation-maximization (EM) algorithm, employed by the research team to illustrate the convergence of the disentanglement process. This convergence is essential for ensuring that the model achieves stable and optimal performance. The EM algorithm provides a structured iterative approach that refines the parameters governing the disentangled representations, thereby optimizing the model’s predictive capabilities.
In addition to the technical advancements embodied in D2-GCN, the researchers introduced an information entropy-based evaluation metric. This innovative metric captures the speed at which the disentanglement process converges. By quantifying this convergence, the team has provided a framework that accentuates the efficiency of D2-GCN compared to its traditional counterparts, paving the way for better understanding of the training dynamics involved in the graph convolutional processes.
The experimental results underscore the efficacy of D2-GCN. The model reported remarkable advancements over conventional methods in both single- and multi-label node classification tasks. With a noticeable improvement in test accuracies, D2-GCN proves to be a formidable contender in the realm of graph-based learning. Additionally, visualizations derived from the model highlight its ability to establish clearer classification boundaries and exhibit higher intra-class similarities. This aspect is particularly noteworthy, as it translates to improved interpretability and reliability when deploying the model in practical scenarios.
Looking forward, the research team envisions integrating subgraph theory into D2-GCN for even greater refinements. By harnessing the potential of subgraph analysis, the model could achieve enhanced local-global disentanglement and a firmer grasp on long-distance dependencies within graphs. Such advancements are crucial, given the intricate relationships typically present in real-world data, whether in social networks, biological systems, or complex organizational structures.
The significance of the work published in the recent study extends beyond mere technical improvements; it illustrates a paradigm shift in how we approach the representation of nodes within graphs. Traditional methods often falter when faced with nuances in neighbor influences, leading to oversimplified characterizations that can misguide predictive outcomes. D2-GCN offers a fresh perspective, emphasizing the importance of dynamic adaptability in modeling node behaviors, ultimately resulting in more refined and accurate machine learning outcomes.
The rigorous methodology applied in the study provides a strong foundation for potential future endeavors in the field of graph neural networks. As researchers continue to explore the depth of graph representations, the innovative ideas reflected in D2-GCN and its architectural frameworks will undoubtedly contribute to advancing our understanding and utilization of complex graphs.
In conclusion, D2-GCN paves the way for a new era in graph neural networks. By resolving the limitations posed by traditional GCNs and introducing dynamic adaptiveness into feature representation, this model stands as a testament to the ingenuity of modern computational methods. With promising results showcased through experimental studies, D2-GCN is set to make waves in various applications—ranging from recommendation systems to social network analysis, and beyond.
The research led by Chuliang Weng is certainly marking a profound advance in the field, igniting excitement about the future of graph-based machine learning methodologies. With each innovation, the potential to transform diverse domains expands, demonstrating that the future of GCNs may be defined by adaptability, precision, and deeper insight into the intricate webs of relationships that define complex systems.
—
Subject of Research: Dynamic Disentangled Graph Convolutional Networks
Article Title: D2-GCN: a graph convolutional network with dynamic disentanglement for node classification
News Publication Date: 15-Jan-2025
Web References: https://journal.hep.com.cn/fcs/EN/10.1007/s11704-023-3339-7
References: 10.1007/s11704-023-3339-7
Image Credits: Shangwei WU, Yingtong XIONG, Hui LIANG, Chuliang WENG
Keywords
Applied sciences, Computer science, Graph neural networks, Node classification, Dynamic models, Machine learning
Tags: adaptive feature channel selectionadvancements in machine learning for graphsD2-GCN for node classificationDynamic Disentangled Graph Convolutional Networkenhancements in graph neural networksepoch-level and layer-level disentanglementevolving graph structures in GCNshyper-adaptive node representationsinnovative approaches to node classification.limitations of traditional GCNsprecise node representation techniquestwo-level disentangling mechanism