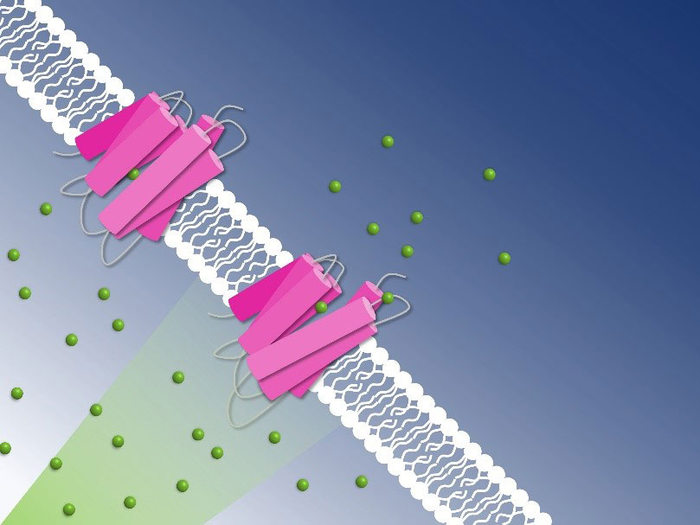
Many bacteria and unicellular algae have light-driven pumps in their cell membranes: proteins that change shape when exposed to photons such that they can transport charged atoms in or out of the cell. Thanks to these pumps, their unicellular owners can adjust to the environment’s pH value or salinity.
Credit: Guillaume Gotthard and Sandra Mous
Many bacteria and unicellular algae have light-driven pumps in their cell membranes: proteins that change shape when exposed to photons such that they can transport charged atoms in or out of the cell. Thanks to these pumps, their unicellular owners can adjust to the environment’s pH value or salinity.
One such bacteria is Nonlabens marinus, first discovered in 2012 in the Pacific Ocean. Among others, it possesses a rhodopsin protein in its cell membrane which transports chloride anions from outside the cell to its inside. Just like in the human eye, a retinal molecule bound to the protein isomerizes when exposed to light. This isomerization starts the pumping process. Researchers now gained detailed insight into how the chloride pump in Nonlabens marinus works.
The study was led by Przemyslaw Nogly, once a postdoc at PSI and now an Ambizione Fellow and Group Leader at ETH Zürich. With his team, he combined experiments at two of PSI’s large-scale research facilities, the Swiss Light Source SLS and the X-ray free-electron laser SwissFEL. Slower dynamics in the millisecond-range were investigated via time-resolved serial crystallography at SLS while faster, up to picosecond, events were captured at SwissFEL – then both sets of data were put together.
“In one paper, we exploit the advantages of two state-of-the-art facilities to tell the full story of this chloride pump,” Nogly says. Jörg Standfuss, co-author of the study who built up a PSI team dedicated to creating such molecular movies, adds: “This combination enables first-class biological research as would only be possible at very few other places in the world beside PSI.”
No backflow
As the study has revealed, the chloride anion is attracted by a positively charged patch of the rhodopsin protein in Nonlabens marinus’ cell membrane. Here, the anion enters the protein and finally binds to a positive charge at the retinal molecule inside. When retinal isomerizes due to light exposure and flips over, it drags the chloride anion along and thus transports it a bit further inside the protein. “This is how light energy is directly converted into kinetic energy, triggering the very first step of the ion transport,” Sandra Mous says, a PhD student in Nogly’s group and first author of the paper.
Being on the other side of the retinal molecule now, the chloride ion has reached a point of no return. From here, it goes only further inside the cell. An amino acid helix also relaxes when chloride moves along, additionally obstructing the passage back outside. “During the transport, two molecular gates thus make sure that chloride only moves in one direction: inside,” Nogly says. One pumping process in total takes about 100 milliseconds.
Two years ago, Jörg Standfuss, Przemyslaw Nogly and their team unravelled the mechanism of another light-driven bacterial pump: the sodium pump of Krokinobacter eikastus. Researchers are eager to discover the details of light-driven pumps because these proteins are valuable optogenetic tools: genetically engineered into mammalian neurons, they make it possible to control the neurons activities by light and thus research their function.
Text: Brigitte Osterath
Journal
Science
DOI
10.1126/science.abj6663
Method of Research
Experimental study
Subject of Research
Cells
Article Title
Dynamics and mechanism of a light-driven chloride pump
Article Publication Date
3-Feb-2022




