Credit: Purdue University/Aritra Bose
In recent years, the market for direct-to-consumer genetic testing has exploded. The number of people who used at-home DNA tests more than doubled in 2017, most of them in the U.S. About 1 in 25 American adults now know where their ancestors came from, thanks to companies like AncestryDNA and 23andMe.
As the tests become more popular, these companies are grappling with how to store all the accumulating data and how to process results quickly. A new tool called TeraPCA, created by researchers at Purdue University, is now available to help. The results were published in the journal Bioinformatics.
Despite people’s many physical differences (determined by factors like ethnicity, sex or lineage), any two humans are about 99 percent the same genetically. The most common type of genetic variation, which contribute to the 1% that makes us different, are called single nucleotide polymorphisms, or SNPs (pronounced “snips”).
SNPs occur nearly once in every 1,000 nucleotides, which means there are about 4 to 5 million SNPs in every person’s genome. That’s a lot of data to keep track of for even one person, but doing the same for thousands or millions of people is a real challenge.
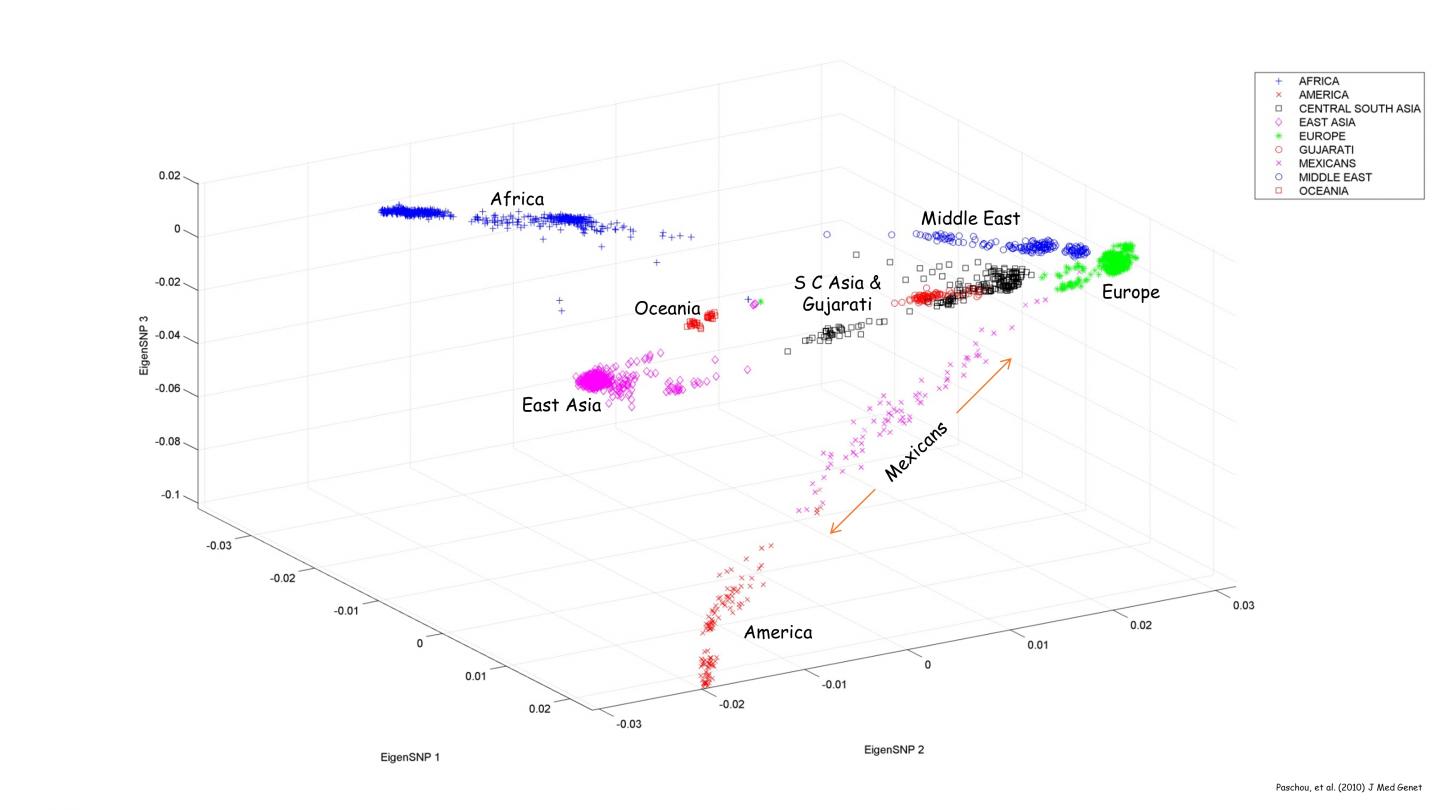
Most studies of population structure in human genetics use a tool called Principal Component Analysis (PCA), which analyzes a huge set of variables and reduces it to a smaller set that still contains most of the same information. The reduced set of variables, known as principal factors, are much easier to analyze and interpret.
Typically, the data to be analyzed is stored in the system memory, but as datasets get bigger, running PCA becomes infeasible due to the computation overhead and researchers need to use external applications. For the largest genetic testing companies, storing data is not only expensive and technologically challenging, but comes with privacy concerns. The companies have a responsibility to protect the extremely detailed and personal health data of thousands of people, and storing it all on their hard drives could make them an attractive target for hackers.
Like other out-of-core algorithms, TeraPCA was designed to process data too large to fit on a computer’s main memory at one time. It makes sense of large datasets by reading small chunks of it at a time.
“In 2017, I met some people from the big genetic testing companies and I asked them what they were doing to run PCA. They were using FlashPCA2, which is the industry standard, but they weren’t happy with how long it was taking,” said Aritra Bose, a Ph.D. candidate in computer science at Purdue. “To run PCA on the genetic data of a million individuals and as many SNPs with FlashPCA2 would take a couple of days. It can be done with TeraPCA in five or six hours.”
The new program cuts down on time by making approximations of the top principal components. Rounding to three or four decimal places yields results just as accurate as the original numbers would, Bose said.
“People who work in genetics don’t need 16 digits of precision – that won’t help the practitioners,” he said. “They need only three to four. If you can reduce it to that, then you can probably get your results pretty fast.”
Timing for TeraPCA also was improved by making use of several threads of computation, known as “multithreading.” A thread is sort of like a worker on an assembly line; if the process is the manager, the threads are hardworking employees. Those employees rely on the same dataset, but they execute their own stacks.
Today, most universities and large companies have multithreading architectures, but FlashPCA2 doesn’t leverage it. For tasks like analyzing genetic data, Bose thinks that’s a missed opportunity.
“We thought we should build something that leverages the multithreading architecture that exists right now, and our method scales really well,” he said. “TeraPCA scales linearly with the number of threads you have. FlashPCA2 doesn’t do this, which means it would take very long to reach your desired accuracy.”
Compared to FlashPCA2, TeraPCA performs similarly or better on a single thread and significantly better with multithreading, according to the paper. The code is available now on GitHub.
###
This research was supported by the National Science Foundation. Vassilis Kalantzis, a Herman H. Goldstine Memorial Postdoctoral Fellow at IBM Research, is a co-first author of the paper.
Media Contact
Kayla Zacharias
[email protected]
Original Source
https:/
Related Journal Article
http://dx.