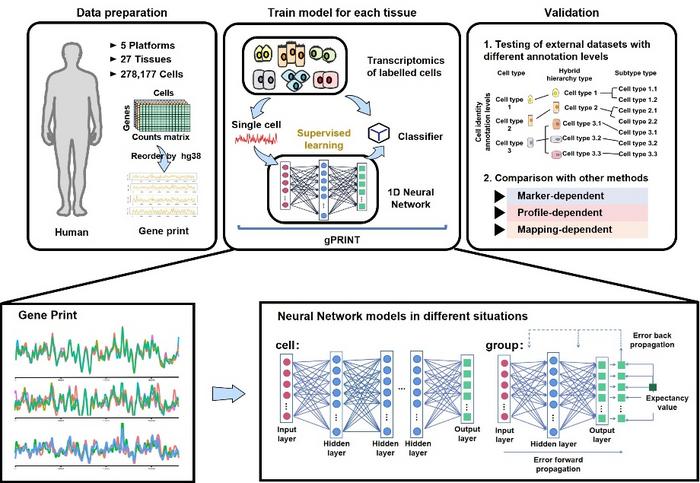
A groundbreaking computational framework named gPRINT has emerged, redefining the landscape of single-cell data integration and disease-specific cell subtype annotation. This innovative approach harnesses the synergy between gene expression profiles and chromosomal positional information to create distinct “gene prints,” an idea inspired by the complex principles underlying speech recognition. By mapping spatial gene organization—particularly the co-regulated gene clusters residing within chromatin structures—gPRINT significantly reduces noise in heterogeneous datasets while enhancing the resolution needed to discern subtle cellular differences.
What sets gPRINT apart from traditional methods is its ability to integrate two critical layers of biological information. Traditional single-cell annotation tools primarily rely on gene expression data alone, which can be confounded by technical artifacts and biological variability. gPRINT transcends these limitations by embedding the spatial context of gene loci, allowing the algorithm to recognize patterns formed by groups of genes physically co-localized on chromosomes. This fusion of transcriptomic and genomic topology data mimics the multidimensional processing seen in human speech recognition technologies, enabling more precise cellular classification.
The power of gPRINT has been demonstrated on an unprecedented scale, with validation conducted over 1.2 million single-cell profiles spanning human tissues collected from multiple public datasets and platforms. By training a neural network on such a diverse dataset, gPRINT achieved a remarkable cross-platform annotation accuracy exceeding 98%. Compared to popular tools like SingleR and Seurat—which depend heavily on either marker genes or clustering—gPRINT excels at resolving ambiguous cellular populations that often confound other methods. For instance, the tumor-stroma interface, an ecotone rich in phenotypic plasticity and cellular intermixing, was delineated with unprecedented clarity using gPRINT.
One of the most striking validations of gPRINT’s utility was its application in tendinopathy research. Here, it uncovered a novel subset of chondrogenic tendon cells marked by co-expression of SOX9 and COL2A1, a population previously undetectable by conventional clustering algorithms or marker-based approaches. This discovery opens new avenues for understanding tendon pathology and potentially designing targeted therapies, underscoring gPRINT’s capacity to illuminate previously hidden cellular players in disease contexts.
Beyond mere annotation, the study elucidates a fundamental mechanistic link between gene prints and three-dimensional genome architecture. Using high-resolution Hi-C chromatin conformation data, researchers confirmed that co-expressed signature genes tend to cluster spatially within the nucleus in disease-specific cell subtypes (DSCSs). For example, clusters such as COL1A1 and ACTA2 on chromosome 7 physically co-localize, reflecting regulatory domains that orchestrate coordinated gene expression. Intriguingly, experimental perturbations disrupting this chromosomal topology—like deletions of key CTCF anchor sites—led to a steep 63% drop in gPRINT’s annotation accuracy. Moreover, CRISPR-mediated enhancer excisions abolished subtype-specific signaling pathways such as TGF-β, underscoring the functional importance of spatial genome organization in maintaining cellular identity.
This three-dimensional perspective transforms our understanding of single-cell heterogeneity. It posits that the genome’s spatial folding patterns are not mere architectural epiphenomena but are integral to the regulatory networks defining disease subtypes. gPRINT leverages this insight to create annotations that are biologically principled and robust across datasets, platforms, and even species.
In a therapeutic context, gPRINT’s integrative database cross-referencing has already yielded promising drug candidates. By interfacing with the Connectivity Map (CMAP) database, researchers prioritized agents like ascorbic acid and celastrol, which exhibit potential to modulate fibrotic pathways characterized by the identified cell subtypes. Such computational drug repositioning efforts could accelerate the development of treatments for fibrosis and related degenerative conditions, wherein cellular heterogeneity and plasticity have complicated conventional therapeutic strategies.
Importantly, the generalizability of gPRINT was highlighted through cross-species validations involving humans, mice, and non-human primates. Conserved fibroblast subpopulations implicated in fibrotic cascades appeared consistently across these models, reinforcing the universality of gene print signatures. This evolutionary conservation affords researchers a powerful translational bridge from animal models to human disease, enhancing the predictive value of preclinical studies.
Application of gPRINT to multi-omics databases such as TendonBase heralds a new era for integrative biomedical research. By unifying transcriptomic, spatial genomic, and epigenomic data under a cohesive analytical framework, gPRINT enables comprehensive decoding of cellular heterogeneity in complex diseases such as fibrosis, cancer, and degenerative disorders. This holistic view promises to unravel pathophysiological mechanisms at unprecedented resolution.
With a robust training set spanning over 159,000 human single cells collected from 26 distinct tissue types and analyzed through five different technological platforms, gPRINT exemplifies the power of big data in biology. Each cell’s gene print is generated by capturing its unique spatially-informed gene expression signature, then classified through a supervised neural network model. Validation on an external dataset further confirmed gPRINT’s superior performance across the hierarchical levels of cell type, hybrid hierarchy type, and traditional subtype classifications.
In summary, gPRINT represents a paradigm shift in single-cell biology. By marrying gene expression with chromosomal spatial information, it delivers a powerful and scalable tool that resolves intra-tissue heterogeneity and discovers novel pathological cellular subpopulations previously inaccessible by conventional means. This breakthrough paves the way for more precise disease modeling, biomarker discovery, and therapeutic targeting in the era of personalized medicine.
This innovative work is detailed in the article titled “Gene print-based cell subtypes annotation of human disease across heterogeneous datasets with gPRINT,” published in Protein & Cell on March 14, 2025 (DOI: 10.1093/procel/pwaf001). As we continue to explore the complex interplay between genome architecture and gene expression, gPRINT stands at the forefront of computational biology, promising to reshape our understanding of cellular identity within health and disease.
Subject of Research: People
Article Title: Gene print-based cell subtypes annotation of human disease across heterogeneous datasets with gPRINT
News Publication Date: 14-Mar-2025
Web References:
https://doi.org/10.1093/procel/pwaf001
Protein & Cell Journal
References:
X Yan R, Fan C, Gu S, Wang T, Yin Z, Chen X. Gene print-based cell subtypes annotation of human disease across heterogeneous datasets with gPRINT. Protein & Cell. 2025 Mar 14; DOI: 10.1093/procel/pwaf001.
Image Credits: Rong Xie, Higher Education Press
Keywords: Cell biology
Tags: cellular classification techniqueschromosomal positional informationco-regulated gene clustersdisease-specific cell subtype annotationgene expression profilesgPRINT computational frameworkinnovative approaches in biomedical researchneural network training in biologynoise reduction in datasetssingle-cell data integrationspatial gene organizationtranscriptomic and genomic topology