Researchers are making strides in the field of gene editing, particularly through techniques like base editing and prime editing. These revolutionary technologies have the potential to directly correct pathogenic mutations in living organisms, thus presenting exciting new avenues for therapeutic applications. However, for these technologies to fulfill their promise, it is vital to accurately measure gene editing events in situ, especially with high spatial resolution. This begs the question of how we can better visualize and quantify these editing events in real-time within native tissues.
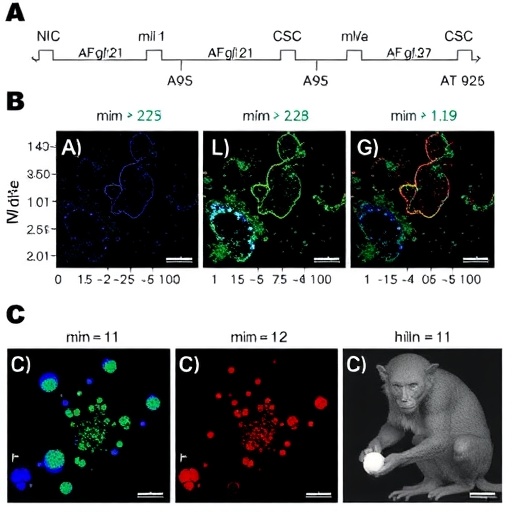
A recent study employed imaging-based in situ sequencing (ISS) to map occurrences of base and prime editing in various tissues of living organisms. This technique holds significant potential for enhancing our understanding of gene editing events in various contexts, including both dividing and non-dividing cells, which are crucial for a range of therapeutic applications. The innovative approach provides an unprecedented ability to pinpoint the exact location and frequency of genomic alterations induced by these groundbreaking editing technologies.
In an impressive display of the technology’s capacity, the researchers utilized ISS in mouse brains treated with intein-split adenine base editors and prime editors delivered through adeno-associated viral vectors. The results provided not only confirmation of the editors’ effectiveness but also rich spatial information that can be pivotal for future advancements. The utilization of viral vectors for delivery is particularly relevant for achieving targeted and efficient gene editing within specific tissues, marking a significant step forward in therapeutic gene editing.
The study further explored the efficacy of base editing technology in the livers of both mice and macaques, treated using adenine base editors encoded on lipid nanoparticle-encapsulated mRNA and guide RNA (RNA-LNP). The outcomes were promising, as effective gene editing was observed across all metabolic zones of liver lobules, indicating a broad distribution of editing events. This also reflects the technology’s ability to penetrate through complex biological environments and reach target cells successfully.
One noteworthy aspect of the research was the testing of repeated doses of RNA-LNP. The initial findings highlighted that the first dose does not adversely influence the editing efficiency or the distribution of subsequent doses. This aspect is particularly reassuring for developing treatment regimens that may require multiple administrations over time. The implications for treating metabolic liver diseases are profound, suggesting that a sustained and effective therapeutic strategy could be established.
The findings demonstrated how ISS can serve as a powerful tool for visualizing and quantifying gene editing events in vivo. This capability could revolutionize the field of gene editing by facilitating real-time assessments of editing efficacy and providing insights into the dynamics of gene modification over time. The importance of such a platform cannot be understated, especially in the context of evaluating novel therapeutic strategies aimed at a variety of genetic disorders.
Another critical point raised by this study is the versatility of RNA-LNPs as delivery mechanisms for gene editing technologies. The ability to encapsulate both mRNA encoding for editors and guide RNA within lipid nanoparticles not only promotes enhanced stability but also fosters efficient cellular uptake. The design of such a delivery system is crucial for achieving the levels of precision required for effective gene editing while minimizing potential off-target effects.
The ramifications of this research extend beyond the confines of academic debate; they signal new hope for patients suffering from genetic disorders and metabolic liver diseases, which often lack effective treatment options. By precisely correcting mutations at the DNA level, the potential for curing diseases traditionally deemed untreatable is becoming increasingly tangible. The seamless fusion of cutting-edge technology with practical applications is poised to change the landscape of gene therapy.
Moreover, the researchers are not only content with their current findings; they are encouraging broader applications of their methodology and results. By laying the groundwork for further exploration of spatial profiling in other tissues and organisms, there is a path forward toward enhancing our arsenal against genetic diseases. The adaptability of ISS could facilitate similar studies in various biological contexts, which would yield additional insights into the complexities of gene editing.
The study’s validation in distinct biological settings fortifies the foundation upon which future developments can be built. As gene editing continues to mature as a discipline, the foundational tools for assessing effectiveness and safety will undoubtedly play a crucial role in its evolution. The researchers believe that continued collaboration between multiple scientific disciplines, including molecular biology, bioengineering, and clinical medicine, will catalyze future breakthroughs.
In conclusion, leveraging advanced imaging technologies like ISS in conjunction with innovative delivery systems such as RNA-LNP reviews the very essence of what is possible in gene editing. The findings from this study represent a promising leap forward, cementing the potential impact of precise genome modifications across a spectrum of therapeutic areas. As the field moves into an era where gene editing may soon arise as a standard practice in clinical settings, the need for thorough validation and a deeper understanding of spatial gene editing dynamics will remain paramount, setting the stage for transformative health outcomes.
As researchers continue to decode the complexities of gene therapy with technologies like base editing and prime editing, the intricate dance between innovation, application, and ethical considerations will shape the future trajectory of the field. It is an exhilarating time for molecular medicine, with the horizon brimming with possibilities that nature previously kept hidden but are now within our grasp.
Subject of Research: Gene Editing Technologies and Their Applications
Article Title: Spatial profiling of gene editing by in situ sequencing in mice and macaques.
Article References:
Janjuha, S., Haenggi, T., Chamberlain, T.C. et al. Spatial profiling of gene editing by in situ sequencing in mice and macaques. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01512-7
Image Credits: AI Generated
DOI: 10.1038/s41551-025-01512-7
Keywords: Gene Editing, Base Editing, Prime Editing, In Situ Sequencing, RNA-LNP, Therapeutic Potential, Metabolic Diseases.
Tags: adenine base editorsadeno-associated viral vectors in gene therapybase editing techniquesgene editing technologiesgenomic alterations mappingimaging-based gene editingin situ sequencing applicationsmouse model gene editingprime editing advancementsreal-time gene editing visualizationspatial resolution in gene editingtherapeutic gene editing