Researchers at Umeå University in Sweden are first to discover that bacteria can multiply disease-inducing genes which are needed to rapidly cause infection. The results were published in Science on June 30, 2016.
More than 22 years ago, researchers at Umeå University were first to discover an infection strategy of human pathogenic Yersinia bacteria — a protein structure in bacterial cell-walls that resembled a syringe. The structure, named "Type III secretion system" or T3SS, makes it possible to transfer bacterial proteins into the host cell and destroy its metabolism.
After the discovery, researchers have found T3SS in several other bacteria species and T3SS has proven to be a common infection mechanism that pathogens, i.e. an infectious agent such as a virus or bacterium, use to destroy host cells. Now, Umeå researchers are again first to find a link between infection and rapid production of the essential proteins needed to form "the poisonous syringe".
Together with researchers at the Helmholtz Centre for Infection Research in Braunschweig, Germany, the Umeå researchers investigated the virulence strategy of Yersinia pseudotuberculosis. This bacterium can cause acute diarrhoea, vomiting and stomach pains, and is closely related to the deadly plague bacterium, which it shares many infection mechanisms with. The genes that these bacteria need for infection are located on a circular extra chromosome, called the virulence plasmid.
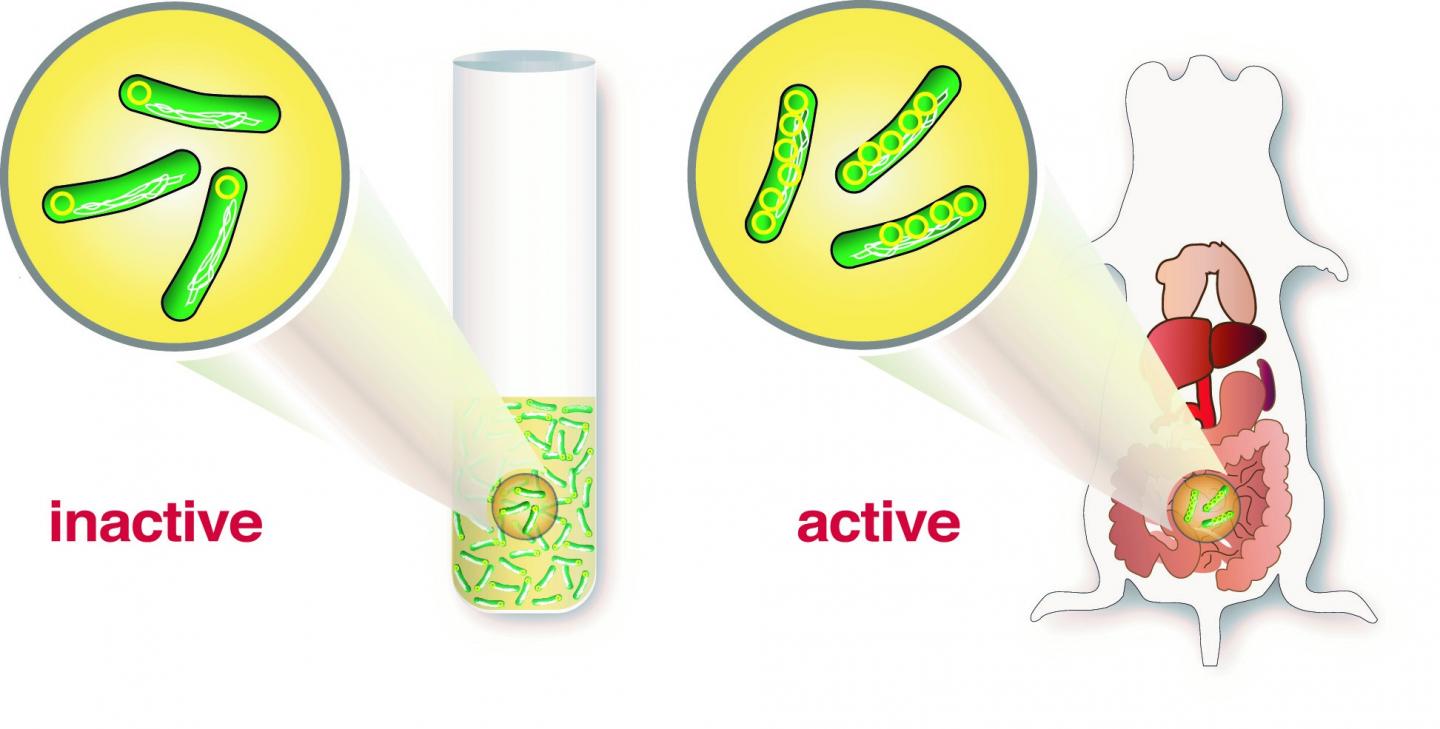
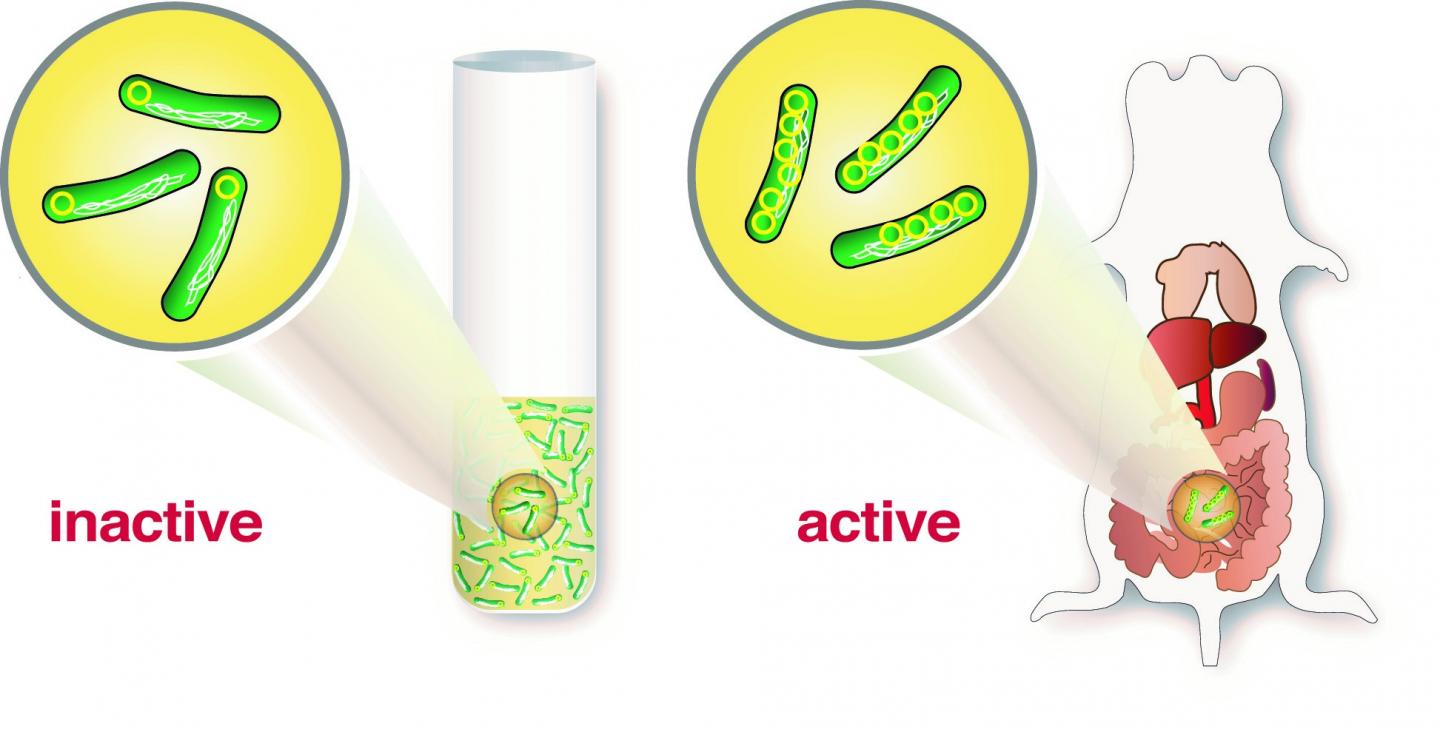
Researchers at Umeå Centre for Microbial Research (UCMR), The Laboratory for Molecular Infection Medicine Sweden (MIMS) at the Department of Molecular Biology first performed infection experiments in cell cultures with human cells and then confirmed their findings using animal models. It turned out that a single copy of the virulence plasmid was not sufficient to induce infection, but the researchers discovered that when Yersinia came in contact with host cells, it triggered a "copying machine" that increased the number of plasmids.
"Yersinia has developed a very clever strategy," says postdoctoral fellow Helen Wang who carried out a large portion of the experiments. "To carry a great number of plasmids, the bacteria need a lot of energy and it negatively affects the bacteria's metabolism and growth. But having one copy of the plasmid as a blueprint that can be rapidly amplified in case of infection is a very clever solution. Many copies of the plasmids give bacteria the opportunity to build up many T3SS and all the proteins needed to quickly knock out host cells during an infection," continues Helen Wang.
It is the first time researchers can demonstrate that an increased number of plasmid-encoded genes is necessary for successful infection by pathogenic bacteria.
"Our study represents a breakthrough in which we show that gene-dosage of plasmid-encoded genes is a fast regulatory strategy used by bacteria. This discovery will contribute to increased insights into bacterial resistance to antibiotics, and is a major step forward in our understanding of how bacteria cause disease," says Tomas Edgren, who together with Hans Wolf-Watz, led the study.
###
Media Contact
Anna Lawrence
[email protected]
46-725-522-918
@UmeaUniversity
http://www.umu.se/umu/index_eng.html