Molecular mechanism of the interplay of clock proteins for generating a circadian oscillation
Credit: Koichi Kato
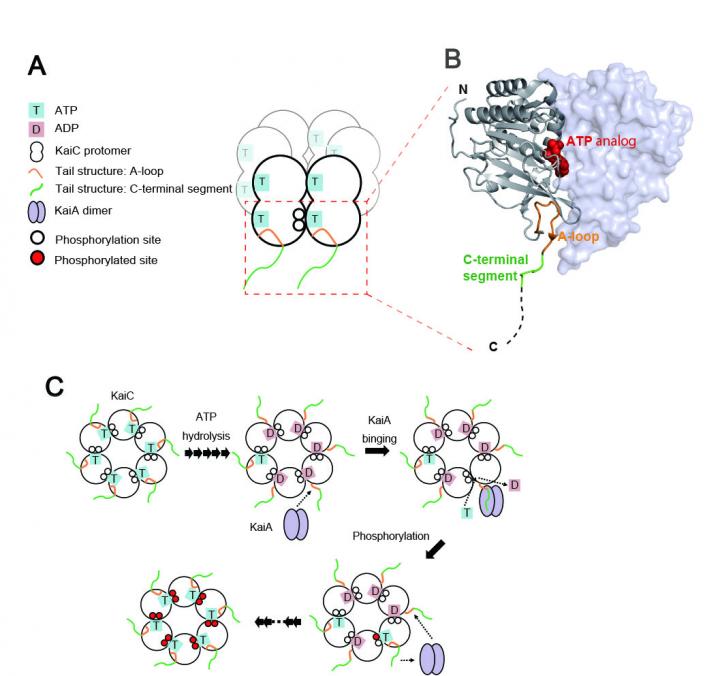
Organisms on this planet, including human beings, exhibit a biological rhythm that repeats about every 24 h to adapt to the daily environmental alteration caused by the rotation of the earth. This circadian rhythm is regulated by a set of biomolecules working as a biological clock. In cyanobacteria (or blue-green algae), the circadian rhythm is controlled by the assembly and disassembly of three clock proteins, namely, KaiA, KaiB, and KaiC. KaiC forms a hexameric-ring structure and plays a central role in the clock oscillator, which works by consuming ATP, the energy currency molecule of the cell. However, it remains unknown how the clock proteins work autonomously for generating the circadian oscillation.
The research groups at Graduate School of Pharmaceutical Sciences of Nagoya City University and Exploratory Research Center on Life and Living Systems (ExCELLS) and Institute for Molecular Science (IMS) of National Institutes of Natural Sciences investigated this mechanism by native mass spectrometry and nuclear magnetic resonance spectroscopy. They found that KaiC degrades ATP into ADP within its ring structure, which triggers the leaping out of the tail of KaiC from the ring. KaiA captures the exposed KaiC tail, facilitating ADP release from the ring, thereby setting the clock ahead.
This “fishing a line” mechanism explains the clockwork interplay of the KaiA and KaiC proteins. Elucidating this mechanism will provide deep insights into not only the circadian clock in cyanobacteria but also that in plants, animals, and humans under physiological and pathological conditions, including jet lag and sleep disorders.
###
Media Contact
Koichi KATO
[email protected]
Original Source
http://www.
Related Journal Article
http://dx.




