Credit: Kinney lab/CSHL, 2019
In this age of “big data,” artificial intelligence (AI) has become a valuable ally for scientists. Machine learning algorithms, for instance, are helping biologists make sense of the dizzying number of molecular signals that control how genes function. But as new algorithms are developed to analyze even more data, they also become more complex and more difficult to interpret. Quantitative biologists Justin B. Kinney and Ammar Tareen have a strategy to design advanced machine learning algorithms that are easier for biologists to understand.
The algorithms are a type of artificial neural network (ANN). Inspired by the way neurons connect and branch in the brain, ANNs are the computational foundations for advanced machine learning. And despite their name, ANNs are not exclusively used to study brains.
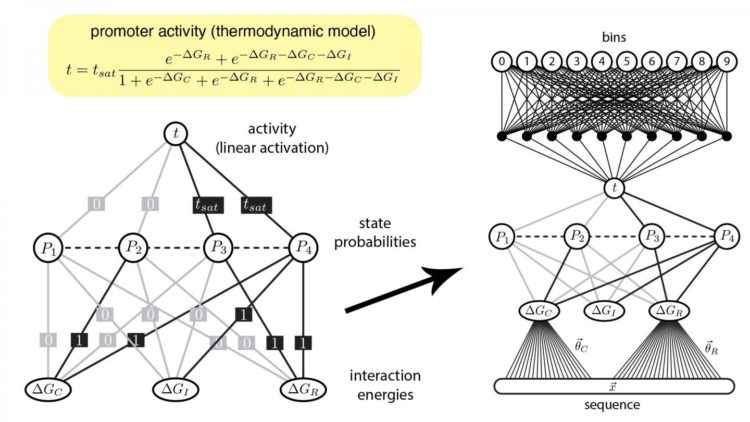
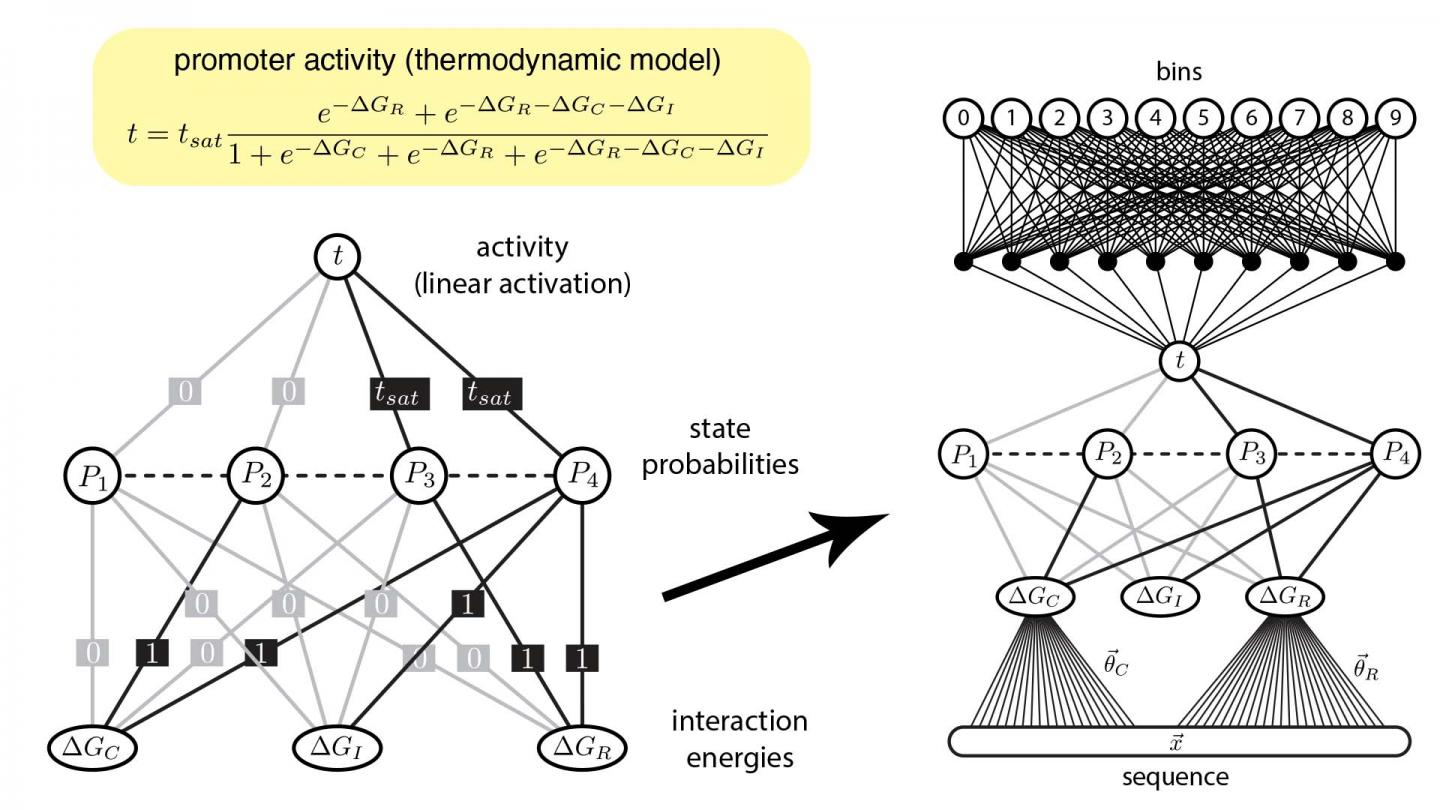
Biologists, like Tareen and Kinney, use ANNs to analyze data from an experimental method called a “massively parallel reporter assay” (MPRA) which investigates DNA. Using this data, quantitative biologists can make ANNs that predict which molecules control specific genes in a process called gene regulation.
Cells don’t need all proteins all the time. Instead, they rely on complex molecular mechanisms to turn the genes that produce proteins on or off, as needed. When those regulations fail, disorder and disease usually follow.
“That mechanistic knowledge–understanding how something like gene regulation works–is very often the difference between being able to develop molecular therapies against diseases, and not being able to,” Kinney said.
Unfortunately the way standard ANNs are shaped from MPRA data is very different from how scientists ask questions in the life sciences. This misalignment means that biologists find it difficult to interpret how gene regulation occurs.
Now, Kinney and Tareen developed a new approach that bridges the gap between computational tools and how biologists think. They created custom ANNs that mathematically reflect common concepts in biology concerning genes and the molecules that control them. In this way, the pair are essentially forcing their machine learning algorithms to process data in a way that a biologist can understand.
These efforts, Kinney explained, highlight how modern, industrial AI technologies can be optimized for use in the life sciences. Having verified this new strategy to make custom ANNs, Kinney’s lab is applying it in investigating a wide variety of biological systems, including key gene circuits involved in human disease.
The results were officially announced in Vancouver, Canada at the 1st Conference on Machine Learning in Computational Biology on December 13. They can be viewed as a preprint on CSHL’s bioRxiv server.
###
Media Contact
Sara Roncero-Menendez
[email protected]
516-367-6866
Original Source
https:/