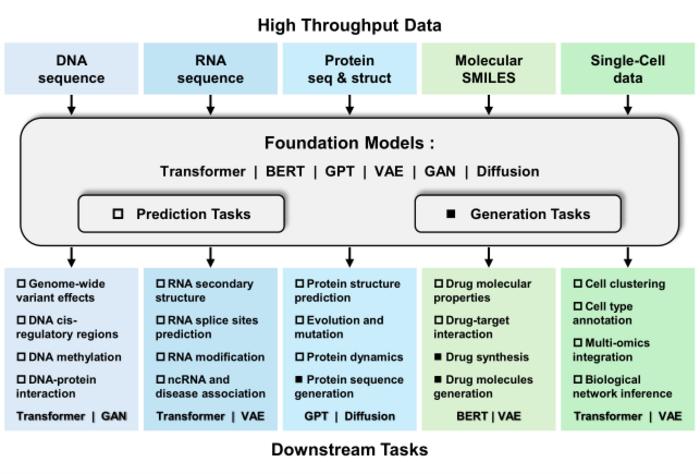
In the rapidly evolving landscape of bioinformatics, researchers are increasingly turning to foundation models (FMs) to harness the power of artificial intelligence in managing and interpreting high-throughput biological data. This recent study led by Prof. Wang and his team at the School of Computer Science and Engineering, Central South University, delves deep into the various advancements made in the integration of FMs within the realm of bioinformatics. By leveraging multiple categories of models, they aim not only to enhance understanding of biological systems but also to equip scientists with the necessary tools to tackle complex biological challenges.
The research identifies four primary categories of foundation models: language, vision, graph, and multimodal FMs. Each category offers unique strengths and capabilities, enhancing the approaches employed in genomics, transcriptomics, proteomics, drug discovery, and single-cell analysis. By systematically categorizing these models, the research provides a roadmap for scientists to select the most appropriate FM based on the specific needs of their bioinformatics applications. As new possibilities unfold, the intersection of AI and molecular biology emerges as a vibrant field ripe for exploration.
At the core of this study lies an emphasis on the versatility and adaptability of foundation models. Prof. Wang and his research team highlighted the potential of these models to be trained using both supervised and unsupervised learning techniques, making them ideal for addressing a spectrum of biological challenges. The research team underscored the necessity of integrating advanced AI technologies into the molecular biology workflow to create a robust framework for future innovations in the field. This integration not only improves predictive capabilities but also allows for a richer analysis of biological phenomena.
Central to this discourse is the understanding of biological databases, training strategies, and hyperparameter configurations, which are crucial components in the deployment of foundation models within bioinformatics. The team meticulously discussed how these attributes could be optimized to enhance performance and accuracy in various tasks. By providing insights into training techniques and configurations, the team lays the groundwork for future research, allowing others to build upon this knowledge and explore new avenues for discovery.
One of the standout contributions of this research is its focus on the evolutionary process of bioinformatics feature mapping. Through a comprehensive understanding of model advancements, the team elucidated how the improved models have mitigated the limitations faced by their predecessors. This evolutionary perspective emphasizes the ongoing nature of research in bioinformatics and highlights the significance of iterative development in crafting effective AI solutions for complex biological inquiries.
The discourse surrounding the practical applications of FMs reached a notable crescendo with the discussion surrounding DeepMind’s efforts in protein structure reconstruction. Prof. Wang pointed to DeepMind’s development of artificial intelligence systems over the past five years, showcasing the promising outcomes that have emerged from these advancements. By tying real-world applications to the evolving field of bioinformatics, the research feeds into a growing narrative of success and innovation, demonstrating the tangible impacts of AI on scientific research.
As the researchers prepared to publish their findings, they captured the broader implications of foundation models in bioinformatics. Their insights into model pre-training frameworks, benchmarking methods, and interpretability promises to shape the future direction of research in the field. The emphasis on model hallucination evaluation reflects a commitment to transparency and reliability, allowing other researchers to engage with these models while understanding their limitations.
Through their work, the team stresses the significance of comprehensive analysis and understanding of the underlying mechanisms that govern biological systems. This approach is essential, as it leads to new insights and generates hypotheses that advance our understanding of complex biological interactions. Additionally, the investigation of how models can interpret and generate biological data serves as a critical component in the continuous effort to bridge gaps in scientific knowledge.
The emerging narrative surrounding foundation models paints a picture of immense potential and possibility, inviting researchers from various domains to converge in interdisciplinary collaborations. By exploring the innovative capabilities of AI and combining expertise from computer science and biology, the research opens doors to novel methodologies that can revolutionize bioinformatics practices and contribute to advances in medicine and health.
Undoubtedly, the burgeoning field of bioinformatics stands at a pivotal juncture, with artificial intelligence-driven solutions poised to redefine biological research paradigms. As the research community rallies around these transformative technologies, the dialogue around foundation models serves as both a catalyst for collaboration and a testament to human ingenuity in the face of scientific challenges.
The team’s final thoughts resonate with a sense of optimism for the future of bioinformatics, as they highlight the potential for foundation models to not only elevate existing practices but also to catalyze groundbreaking discoveries. By charting this course, they aim to inspire a new generation of scientists to leverage AI technology in their quest to understand the intricacies of life itself.
In conclusion, this pivotal study encapsulates the essence of interdisciplinary collaboration and the transformative power of foundation models in bioinformatics. By highlighting theoretical underpinnings and practical applications, the researchers not only contribute to the academic landscape but also underscore the inevitability of AI’s central role in the future of biological research.
Subject of Research: Foundation Models in Bioinformatics
Article Title: Leveraging AI: The Future of Foundation Models in Bioinformatics
News Publication Date: October 2023
Web References: http://dx.doi.org/10.1093/nsr/nwaf028
References: National Science Review
Image Credits: ©Science China Press
Keywords: bioinformatics, foundation models, artificial intelligence, genomics, high-throughput data, molecular biology, protein structure, DeepMind, AI models, interdisciplinary research, biomedical applications.
Tags: AI-driven biological insightsartificial intelligence in biologycategories of foundation modelschallenges in bioinformatics researchfoundation models in bioinformaticsgenomics and transcriptomics applicationshigh-throughput biological data analysisintegration of AI and bioinformaticslanguage and vision models in bioinformaticsmultimodal models in molecular biologyproteomics and drug discoverysingle-cell analysis techniques