Maize, known scientifically as Zea mays, is a staple crop with profound implications for global food security. As one of the most cultivated cereal grains worldwide, maize plays an influential role in agriculture, providing essential nutrients and energy. The evolution of maize breeding has been characterized by significant advancements, particularly through the use of heterosis or hybrid vigor, which enhances yields and resilience in various environmental conditions. However, effectively breeding for elite inbred lines, which are critical for developing hybrid varieties, presents numerous obstacles. Challenges include the complex and diverse genetic architecture of maize, the vast origins of germplasm, and the limitations in predicting phenotypic traits accurately.
Recent innovations in genomic technologies, coupled with artificial intelligence, have paved the way for enhanced understanding of maize genetics. These modern approaches offer a deeper insight into the complexities of genetic variability present within maize populations, significantly aiding in the selection of desirable traits. The application of next-generation sequencing has become crucial in tracking genetic markers that inform breeding programs and help in maximizing the performance of inbred lines. The convergence of genomics and machine learning not only streamlines breeding processes but also opens avenues for improving overall crop health and yield.
A pivotal research effort spearheaded by scientists at the Institute of Crop Science, Chinese Academy of Agricultural Sciences, has made remarkable strides in this field. Their paper, titled “Genomic analysis of modern maize inbred lines reveals diversity and selective breeding effects,” was recently published in the esteemed journal Science China Life Sciences. This comprehensive study investigated a cohort of 2,430 inbred lines derived from elite commercial hybrids distributed across diverse geographical regions. Additionally, another 503 inbred lines were sourced from natural populations, resulting in a well-rounded, genetically diverse inbred population for examination.
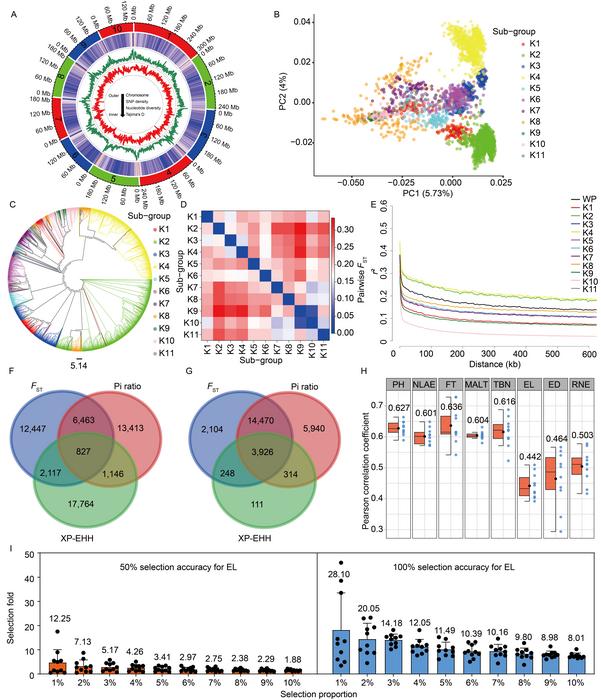
The extensive research included the resequencing of this large-scale maize population, which yielded substantial genomic data. Following stringent quality control measures and variant calling procedures, a remarkable 437,081 high-quality single nucleotide polymorphism (SNP) markers were identified, representing a significant step in the understanding of maize’s complex genetic landscape. These SNPs serve as critical indicators for breeding practices, providing vital information about genetic variation and helping to inform decisions that impact crop performance.
Through rigorous analysis, the researchers shed light on the distribution of genomic variation and explored the nuances of genetic diversity within the maize population. They identified distinct heterotic group types and characterized the population differentiation associated with modern breeding practices. This research illuminated the substantial effects of artificial selection on the maize genome, revealing how selective pressures have shaped its evolution. Importantly, the study identified two potential new heterotic groups, broadening the genetic toolkit available for maize breeding.
The study’s authors conducted a selection sweep analysis that compared the modern inbred lines with those from natural populations. This analysis unveiled multiple loci that are likely under selection during the breeding process, pinpointing genes linked to critical agronomic traits such as flowering time, root development, stress resistance, yield, and plant morphology. These findings emphasize the significance of specific genomic regions that are crucial for optimizing maize performance under varying environmental conditions.
To further leverage their findings, the research team employed advanced deep learning algorithms to develop genomic prediction models aimed at forecasting eight vital traits related to maize plant architecture and yield. These predictive models demonstrated impressive accuracy in predicting target traits, validating the reliability and relevance of the identified loci. Such predictive modeling represents a significant advancement in the capacity to enhance breeding programs, potentially leading to improved crop varieties that can withstand the challenges posed by climate change and growing populations.
Additionally, the researchers introduced an innovative concept known as the selection proportion, aiding in the exploration of the dynamics between the size of validation populations and the efficiency of breeding selection. Through simulation analyses, they revealed that the selection proportion is a pivotal factor influencing genetic gains in yield-related traits. This insight holds potential for revolutionizing breeding strategies, thereby optimizing resource allocation and enhancing genetic progress in maize breeding efforts.
The implications of this research extend beyond immediate genetic applications; they also provide a vision for the future of maize breeding. By establishing a diverse germplasm resource pool and uncovering new heterotic groups, the authors contribute significantly to the genetic repository available for maize improvement initiatives. Meanwhile, the genomic prediction models they developed offer researchers and breeders alike a sophisticated tool to inform breeding decisions effectively.
Collectively, the insights gleaned from this research underscore the importance of integrating genomic information into breeding programs. By harnessing the power of advanced technological tools and large-scale genomic data, maize breeding can become more targeted and effective. As the global demand for food continues to rise, studies like these not only reflect ongoing scientific progress but also illuminate the path towards sustainable agricultural practices that can meet future challenges.
In conclusion, the exploration of maize inbred lines through extensive genomic analysis has yielded substantial advancements in understanding the genetic underpinnings that govern this vital crop. The research illustrates how modern genomics serves as a beacon of hope for enhancing maize breeding efficiency and effectiveness, addressing the pressing challenges of food security and agricultural sustainability. As the scientific community continues to delve into the genetic intricacies of maize, the potential for impactful innovations in crop breeding and enhancement remains immense.
Subject of Research: Genetic analysis of maize inbred lines
Article Title: Genomic analysis of modern maize inbred lines reveals diversity and selective breeding effects
News Publication Date: Not specified
Web References: http://dx.doi.org/10.1007/s11427-024-2725-1
References: Not specified
Image Credits: ©Science China Press
Keywords
Maize, genomic analysis, breeding, SNP markers, heterosis, artificial selection, genetic diversity, deep learning, trait prediction, crop improvement, food security, germplasm resource.