Verifying the efficacy and safety of a drug, clinical trials are a hurdle that must be cleared before a new drug hits the market. Conducting them on humans would be the most ideal but due to stringent ethical standards, patient availability, and limited funding, testing drug efficacy is mostly conducted on animal models. Therefore, it is considered critical to minimize the difference between human and animal disease models to accurately verify the effectiveness of the drug being tested.
Credit: POSTECH
Verifying the efficacy and safety of a drug, clinical trials are a hurdle that must be cleared before a new drug hits the market. Conducting them on humans would be the most ideal but due to stringent ethical standards, patient availability, and limited funding, testing drug efficacy is mostly conducted on animal models. Therefore, it is considered critical to minimize the difference between human and animal disease models to accurately verify the effectiveness of the drug being tested.
Recently, a POSTECH research team led by Professor Sanguk Kim and Ph.D. candidate Doyeon Ha (Department of Life Sciences) has developed a technology to select genes from animal models to accurately mimic human diseases. Recently published in Nucleic Acids Research, an authoritative journal in the field of functional genomics, these findings are anticipated to help develop new drugs and effective therapeutics by engineering more precise animal models.
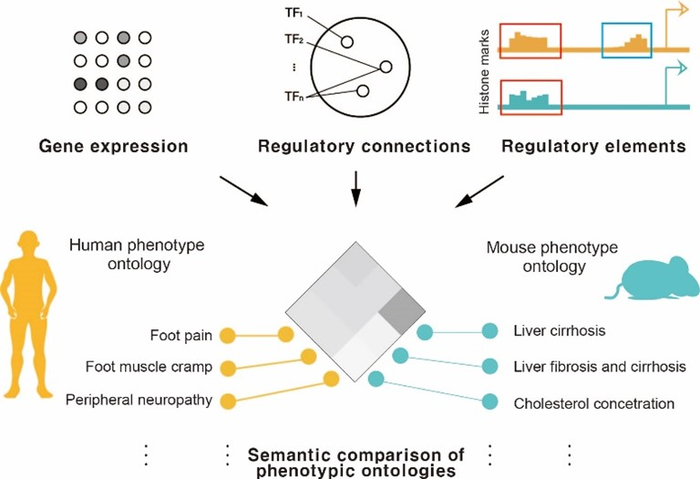
The research team analyzed the aggregate disease symptoms observed in human and mouse models using big data. Unlike conventional research that were conducted based on genomic sequence of orthologous genes across species, this study focused on the tissue-specific gene expression patterns and the gene regulatory networks they form.
Experimental analysis confirms that when the gene regulatory network of two species differ between human and mouse models, the mutation model of the target gene is not suitable for studying the human disease. Furthermore, analyzing the rewiring of the regulatory networks in human and mouse models help select an animal model that mimics the human disease with higher accuracy.
“Using this technology, we can predict the success or failure of animal disease models before making them and help develop more effective new drugs by better understanding disease mechanisms,” remarked Professor Sanguk Kim on the significance of the study.
This study was conducted with the support from the Medical Device Innovation Center and the Graduate School of Artificial Intelligence at POTSECH and from the Mid-career Researcher Program of the National Research Foundation of Korea.
Journal
Nucleic Acids Research
DOI
10.1093/nar/gkac050
Article Title
Evolutionary rewiring of regulatory networks contributes to phenotypic differences between human and mouse orthologous genes
Article Publication Date
7-Feb-2022




