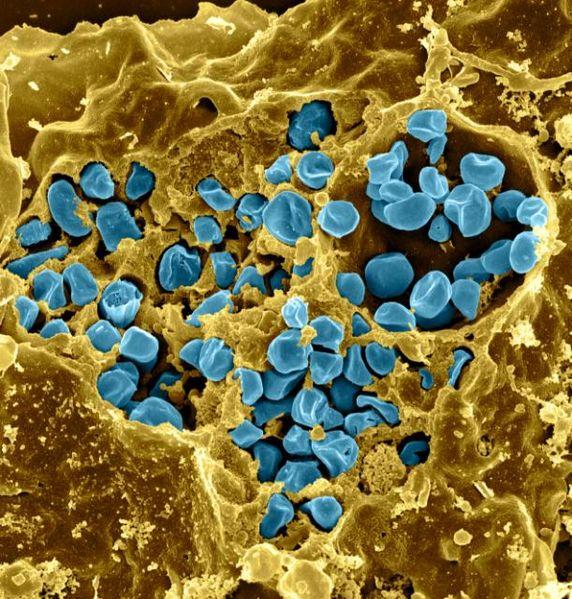
Credit: NIAID
LOS ALAMOS, N.M., Dec. 19, 2016 — The virulent pathogen that causes the disease tularemia, or "rabbit fever," was weaponized during past world wars and is considered a potential bioweapon. Through a new study of the coccobacillus Francisella, Los Alamos National Laboratory researchers are working to use DNA markers to discern related but relatively harmless species as they are identified and to provide a means to distinguish them from the harmful F. tularensis.
"This large study is particularly notable for having used 31 publicly available genomes plus select genes from about 90 additional isolates," said corresponding author Cheryl Kuske of the Bioenergy & Biome Sciences group at Los Alamos National Laboratory. The paper was published in the current issue of Applied and Environmental Microbiology (AEM).
The group's work includes developing a comprehensive, genomics-based understanding of organisms and their environment as part of the Lab's energy-security research into energy sources and their impact. "We conducted standard single and multi-gene comparisons with whole-genome approaches and identified potential virulence factors to discriminate new species from among the more traditional set. Thanks to coordination with the Centers for Disease Control and Prevention (CDC), we now have a framework to identify new isolates or environmental detections even if only partial information is available," Kuske said.
Humans can easily develop tularemia through incidental contact with infected rabbits, cats, rodents or other animals (and the ticks or fleas that may be on them). The disease manifests in a variety of forms: fever and chills with joint pain, cough or pneumonia, or abdominal pain and distress. Fatality rates range, depending on the form of the disease, from 2 percent to 24 percent, according to CDC data. Due to this severity, the CDC has made tularemia a reportable disease, and like the pathogens causing anthrax and plague, it is considered a bioweapon.
Through detailed genome comparisons, sequence-alignment algorithms and other bioinformatics tools run on Los Alamos computers, the Los Alamos team identified features that differentiate among F. tularensis and other novel clinical and environmental Francisella isolates, providing a knowledge base for comparison of new sequences from clinical or environmental surveys.
"For residents of areas such as the Southwest, where tularemia outbreaks are endemic and potentially life threatening, having more identification tools at hand is extremely valuable," Kuske said. The distribution of these isolates is global, however, suggesting a worldwide distribution of Francisella organisms that inhabit fish, ticks and a variety of environmental sources. The team designated four new species groups within the genus; Francisella opportunistica, an opportunistic pathogen of immune compromised patients; Francisella salina and Francisella uliginis, environmental isolates from coastal seawater near Galveston, TX; and Francisella frigiditurris from cooling-tower water in California.
###
The paper: "Whole genome relationships among Francisella bacteria of diverse origin define new species and provide specific regions for detection." (http://aem.asm.org/content/early/2016/11/21/AEM.02589-16.long)
Funding: The genomic sequencing was funded by the U.S. Department of Homeland Security, Science and Technology Directorate.
Media Contact
Nancy Ambrosiano
[email protected]
505-667-0471
@LosAlamosNatLab
http://www.lanl.gov