Credit: Associate Professor Tokio Tani
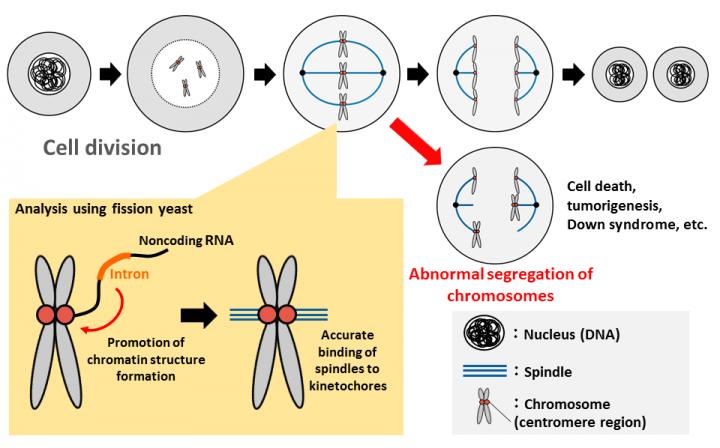
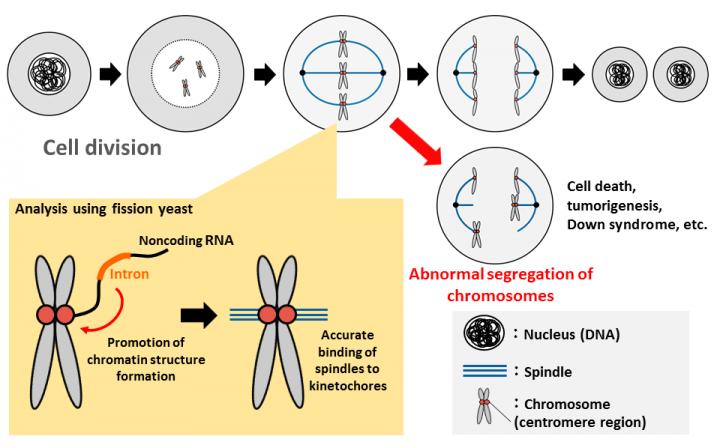
When cells divide, chromosomes need to be evenly segregated between daughter cells. This equal distribution of chromosomes is very important to accurately pass on genetic information to the next generation. Abnormal chromosomal segregation, on the other hand, can cause cell death (apoptosis) or diseases like Down syndrome and cancer. In order for chromosomes to equally divide, it is necessary first to bind the filamentous spindle fiber to a specific region, the centromere, of the chromosome. For the spindle fiber to be correctly joined, it is essential that a part of the chromosome has a special structure called heterochromatin. However, the mechanism for forming this structure has not been sufficiently clarified.
Joint research between Kumamoto University, Osaka University, and the National Institute for Basic Biology in Japan used fission yeast to show that some RNA sequences called "introns" promote heterochromatin structure formation. Since introns are normally excised from RNA after transcription, and do not exist when genes are functioning, it was thought that they were of no particular use. The discovery made by this research, however, revealed that introns have functions that are extremely important to organisms during chromosome segregation. The finding is expected to lead to the clarification of development mechanisms of diseases caused by abnormalities in the number of chromosomes, such as Down syndrome.
The chromosome centromere is an essential area for chromosome movement and accurate segregation in which a kinetochore structure forms a connection to a spindle fiber. Chromosomal DNA of fission yeast and human centromeres form a heterochromatin structure in which the transcription is suppressed in a compactly folded state. The formation of that heterochromatin structure is known to be essential for the kinetochore formation on the chromosome centromere.
The fission yeast used in this analysis is a unicellular eukaryote and is often used for research as a model organism in molecular biology because it is easily genetically manipulated for analysis of mutant strains. Furthermore, 40% of fission yeast genes have introns, compared to about 4% of genes in other yeasts, making it the most similar to humans since most human genes have introns. Interestingly, noncoding RNA (RNA that does not make proteins) in fission yeast transcribed from the chromosome centromere is necessary for the formation of heterochromatin in the centromere itself.
Most genes of higher eukaryotes, such as humans, do not sequentially encode genetic information as is seen in an organism like Escherichia (E.) coli. They are interrupted by introns. These intron sequences are transcribed from DNA to RNA and then excised in the nucleus (splicing reaction). This is why intron sequences have traditionally been regarded as meaningless sequences without genetic information.
Professor Tokio Tani's research group in Kumamoto University found that a mutation in the protein Prp16, which is involved in the splicing reaction that removes intron sequences, makes it impossible to accurately separate chromosomes during cell division. They examined why splicing mutants show chromosome segregation abnormalities and found that splicing mutant strains do not normally form heterochromatin in the chromosome centromere region. They then analyzed the cause of abnormality of centromeric heterochromatin formation in splicing mutant strain, showing that introns in nontranslatable RNA are transcribed from chromosome centromeres, and that artificial removal of introns induced abnormalities in heterochromatin formation. Furthermore, the research group found that splicing factors and heterochromatin-forming factors interact with each other, and that splicing factors gather on introns in noncoding RNA synthesized from the centromere region to form a platform for promoting heterochromatin formation.
This study revealed for the first time that intron sequences, originally thought to be useless sequences that were normally removed in the process of gene expression, have an important function of controlling heterochromatin formation in chromosome centromeres. "We have also found that inhibition of splicing factor expression causes abnormality of chromosome segregation in human cells, and clarified the possibility that a similar mechanism exists in humans," said Professor Tani. "The results of this research are expected to be useful for elucidating the mechanism of the occurrence of diseases caused by abnormalities in the number of chromosomes, such as Down syndrome, and may lead to treatments in the future."
###
This finding was first reported in PLOS Genetics on February 23th, 2017.
[Citation]
M. Mutazono, M. Morita, C. Tsukahara, M. Chinen, S. Nishioka, T. Yumikake, K. Dohke, M. Sakamoto, T. Ideue, J.-i. Nakayama, K. Ishii, and T. Tani, "The intron in centromeric noncoding rna facilitates rnai-mediated formation of heterochromatin." PLoS Genetics, vol. 13, p. e1006606, Feb. 2017. DOI: 10.1371/journal.pgen.1006606
Media Contact
J. Sanderson, N. Fukuda
[email protected]
http://ewww.kumamoto-u.ac.jp/en/news/