Engineering proteins for desirable traits has been the holy grail of modern biotechnology. For example, the food industry can benefit from engineered enzymes which have the ability to enhance biochemical reactions at higher temperatures, as compared to natural enzymes. This can save valuable resources such as labor, money, and time. However, the process of arriving at a functional protein of interest with the desired trait presents significant challenges.
Credit: The authors
Engineering proteins for desirable traits has been the holy grail of modern biotechnology. For example, the food industry can benefit from engineered enzymes which have the ability to enhance biochemical reactions at higher temperatures, as compared to natural enzymes. This can save valuable resources such as labor, money, and time. However, the process of arriving at a functional protein of interest with the desired trait presents significant challenges.
Current protein engineering approaches, such as directed evolution, rely heavily on chance to narrow down ideal variants of the protein of interest. Directed evolution uses repeated introductions of protein sequence alterations called mutations (iterative mutagenesis) followed by quick screening of large numbers of the variant proteins (high-throughput screening). Not surprisingly, this method is labor-intensive and inefficient.
To overcome these limitations, a group of researchers from China led by Dr. Huifeng Jiang from the Tianjin Institute of Industrial Biotechnology at the Chinese Academy of Sciences and National Center of Technology Innovation for Synthetic Biology, developed a protein engineering strategy predicated on artificial intelligence called “DeepEvo.” Explaining further, Dr. Jiang states, “DeepEvo uses a deep evolution strategy, combining principles of deep learning—a process that emulates how the living brain functions—and evolutionary biology.” The study was published online in BioDesign Research on 20 March, 2024.
The researchers applied DeepEvo for engineering high-temperature tolerance in an enzyme called glyceraldehyde-3-phosphate dehydrogenase (G3PDH). G3PDH breaks down glucose to generate energy during glycolysis in living cells. When the team experimentally validated the DeepEvo results, they achieved a promising success rate of over 26%.
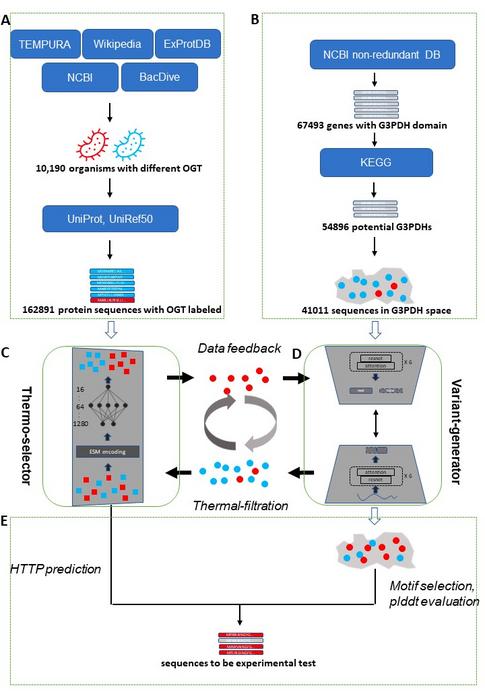
In the study, the data used for DeepEvo involved sequences from organisms with varying optimal growth temperatures (OGT) and naturally occurring sequences with desired functions. The developed DeepEvo strategy comprised a selector (Thermo-selector) and a variant generator (Variant-generator) to yield functional protein sequences incorporating the desired trait. While the selector acted as a selective pressure to enrich the desired protein sequences, the variant generator produced these sequences—in this case, G3PDH variants with high-temperature tolerance. The sequences labeled with OGT trained the Thermo-selector, while those with the desired function trained the Variant-generator. The Thermo-selector filtered sequences, guiding the Variant-generator.
Notably, a protein language model—a type of deep learning model—formed the basis for the Thermo-selector used in this study. Such models are trained on vast amounts of real-world protein sequence data to learn the patterns and features inherent in those sequences. This developed selector uses a learned representation of protein sequences to guide the generation and selection of sequences with the desired trait.
Furthermore, the researchers accumulated high-temperature tolerance traits in the protein sequences through an iterative process involving the generator and selector. Iterative refinement of sequences predicted as high-temperature tolerant formed a cycle of sequence generation. Explaining further, Dr. Jiang adds, “The iterative process involved in DeepEvo mimics the process of natural selection, where functional sequences are favored and accumulated over successive generations, ultimately leading to the development of protein variants with the desired properties.“
Consequently, the researchers verified the predicted high-temperature tolerant protein sequences conserving functional motifs through wet lab experiments. From the 30 generated sequences, they obtained eight variants, thus highlighting the high efficiency of DeepEvo in engineering proteins with high-temperature tolerance.
Going ahead, DeepEvo could aid the selection of diverse traits of interest, not just high-temperature tolerance. In this regard, Dr. Jiang remarks, “We could apply the DeepEvo approach for engineering other protein properties such as acid-base tolerance, catalytic activity, and antigen affinity, facilitating the generation of new proteins with multiple desired properties.”
DeepEvo has thus, paved the way for efficient protein engineering, all thanks to the efforts of Dr. Jiang and his research group. Easy and efficient production of proteins customized for desired traits may soon become a reality.
###
References
DOI
10.34133/bdr.0031
Original Source URL
https://spj.science.org/doi/10.34133/bdr.0031
Authors
Huanyu Chu1,2†, Zhenyang Tian 1,2,3†, Lingling Hu1,2,4, Hejian Zhang 1,2,4,Hong Chang1,2,4, Jie Bai 1,2,4, Dingyu Liu1,2, Lina Lu1,2, Jian Cheng 1,2,and Huifeng Jiang 1,2*
Affications
1 Key Laboratory of Engineering Biology for Low-carbon Manufacturing, Tianjin Institute of IndustrialBiotechnology, Chinese Academy of Sciences, Tianjin 300308, P. R. China.
2 National Center of TechnologyInnovation for Synthetic Biology, Tianjin 300308, P. R. China.
3Tianjin Zhonghe Gene Technology Co., LTD,Tianjin 300308, P. R. China.
4College of Biotechnology, Tianjin University of Science and Technology,Tianjin 300457, P. R. China.
Journal
BioDesign Research
DOI
10.34133/bdr.0031
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
High-temperature Tolerance Protein Engineering through Deep Evolution
Article Publication Date
3-Apr-2024
COI Statement
The authors declare that they have no competing interests.




