The nervous system is made up of diverse cells that arise from progenitors in a specific time-dependent pattern. In a new study, published in Nature Communications, researchers have uncovered the molecular players involved and how the timing is controlled.
Credit: Li lab
The nervous system is made up of diverse cells that arise from progenitors in a specific time-dependent pattern. In a new study, published in Nature Communications, researchers have uncovered the molecular players involved and how the timing is controlled.
“We are interested in studying how neural diversity is created during animal development. First, the stem cells produce different types of neurons, which in turn construct the brain,” said Hailun Zhu, a graduate student in the Li group.
The generation of neural diversity by neural progenitors, called neuroblasts, is regulated in two distinct ways: spatially, where neuroblasts at different locations make different neuron types, and temporally, by which the same neuroblasts generate different neuron types as they age.
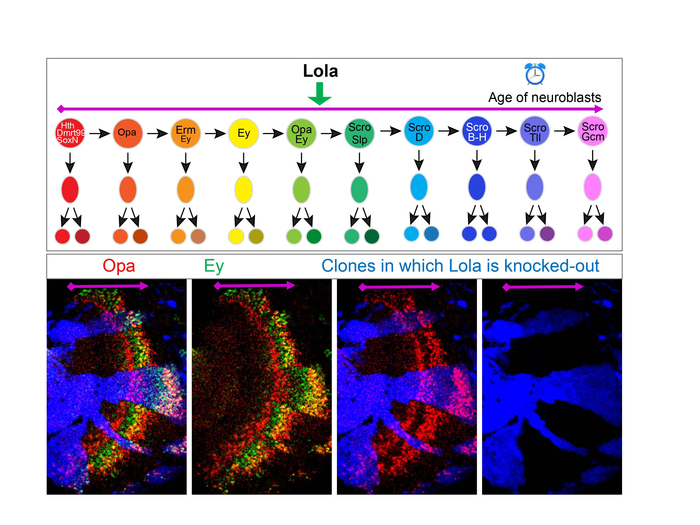
“We are focused on the temporal patterning of neuroblasts, and we use the Drosophila medulla, which is a part of the visual processing center, as a model” said Xin Li (GNDP), an assistant professor of cell and developmental biology.
Li’s postdoctoral work had revealed that there is a cascade of Temporal Transcription Factors in Drosophila medulla neuroblasts where some factors are expressed early on and they successively activate others. However, gaps were observed in this original cascade, and it was also not known how the temporal cascade progression was regulated.
To solve these problems, the Li group, in collaboration with Sihai Dave Zhao (GNDP), an associate professor of statistics, used single-cell RNA sequencing technology to examine how gene expression changes as medulla neuroblasts age.
“We used two markers to label the cells. One was expressed in all the neuroblasts and the other was expressed specifically in the medulla part of the optic center,” Zhu said. “We then sorted the cells and sequenced the RNA in single medulla neuroblasts.”
Single cell RNA sequencing adds specific barcodes to the transcripts that are formed when the information in the DNA is converted to RNA. As a result, each cell has a different barcode and at the end the researchers can identify the transcripts in every single cell. The sequenced cells were then grouped based on the similarity in their gene expression.
The researchers found that the TTFs that had previously been identified by Li’s postdoctoral work were expressed in specific cell clusters. These results demonstrated that medulla neuroblasts were indeed clustered according to their age, from the youngest to the oldest. Furthermore, the researchers identified nine more transcription factors that are expressed in temporal patterns in medulla neuroblasts.
The group confirmed their findings using mutant Drosophila. They found that early transcription factors are required to activate late transcription factors, while late transcription factors repress early ones, forming a temporal cascade. “We used mutants that lacked these transcription factors to test whether they’re required in the cascade. We were able to find some TTFs that were missing in the previous studies, allowing us to develop a more complete temporal patterning gene network,” Zhu said.
They also discovered that the speed of the cascade progression is regulated by transcription factors that are not TTFs. “We found that although the transcription factor Lola is not expressed in a specific stage, without it the cascade slows down,” Zhu said. “It’s very interesting and it’s different from what we’ve seen before.”
The researchers are interested in further examining the transcriptional mechanisms regulating the TTF network. “The interactions between the transcription factors inferred from the mutant analysis are not necessarily direct. The next step is to see whether they act directly,” Li said. “We also want to examine how these TTFs control the downstream specification of different neuron types.”
The study “A comprehensive temporal patterning network in Drosophila medulla neuroblasts revealed by single-cell RNA sequencing” was published in Nature Communications and can be found at https://www.nature.com/articles/s41467-022-28915-3. The work was funded by NSF-Simons Center for Quantitative Biology at Northwestern University and the National Eye Institute.
Journal
Nature Communications
DOI
10.1038/s41467-022-28915-3
Article Title
A comprehensive temporal patterning gene network in Drosophila medulla neuroblasts revealed by single-cell RNA sequencing.
Article Publication Date
10-Mar-2022




