‘Hidden’ associations between the human virome and human health and disease
Credit: ©Kei Sato
Human-resident microbes can influence both health and disease. Investigating the microbiome using next-generation sequencing(*1) technology has revealed examples of mutualism and conflict between microbes and humans.
Compared to bacteria, the viral component of the microbiome (i.e. the “virome”) (*2) is understudied. Somatic tissues of healthy individuals are usually inaccessible for virome sampling. Therefore, there is limited understanding of the presence and distribution of viruses in tissues in healthy individuals and how virus infection associates with human gene expression and perturbs immunological homeostasis.
A research group at The Institute of Medical Science, The University of Tokyo (IMSUT) comprehensively described the “human tissue virome atlas” using a public database.
“The atlas we described will be useful to address the currently “hidden” associations between the human virome and human health and disease. Moreover, our analytic pipeline is versatile, and therefore, will be applicable to a variety of virological studies including the current COVID-19 pandemic.”, said the lead scientist, Kei Sato, Associate Professor (Principal Investigator) in the Division of Systems Virology, Department of Infectious Disease Control, IMSUT.
Microbiome in the era of next-generation sequencing (NGS)
Advances in NGS methods in recent decades have made comprehensive surveys of a variety of microorganisms possible. Metagenomic analyses(*3) have explored microorganisms, including bacteria, phages, and viruses, in a variety of places on Earth, such as the oceans and soils. Perhaps the most deeply surveyed microbiome is that of humans.
The Human Microbiome Project aims to characterize bacteria, viruses, and other microorganisms in the human body. As they often live outside human cells, the human bacterial microbiome has been well described in multiple organs and samples via non-destructive sampling: the bacterial microbiome of the skin, oral cavity, and gastrointestinal tract (including feces) are well described. A major theme emerging from these studies is that, while certain microbial species are associated with pathology, many components of the microbiome likely play a symbiotic role in maintaining human health.
What is the “virome”
Many viruses are clearly human pathogens: HIV, influenza virus and a novel coronavirus (designated as SARS-CoV-2) are the causative agents of human diseases. On the other hand, similar to the case of the human bacterial microbiome, some viruses can chronically infect a broad range of human tissues without overt pathology. Previous studies have suggested that these viruses nonetheless have detrimental effects. For example, the human respiratory syncytial virus and human rhinoviruses may play important roles in the inception of childhood asthma and atopic asthma, respectively.
On the other hand, there are a few examples of viruses that exhibit protective effects. For example, GB virus C infection can be protective against HIV infection and improve survival. Thus, virus infections are associated with multiple aspects of human health, and so revealing the human “virome” would be beneficial for understanding the hidden mutualism and/or conflict between humans and viruses. Previous studies of the human virome have used specimens that are relatively easy to access in healthy individuals (e.g., blood or skin).
In other words, it is technically difficult to obtain the somatic tissues from inside the human body (e.g., brain and internal organs) of healthy individuals for virome investigation. Moreover, it remains unclear 1) what kinds of viruses infect various tissues in healthy individuals and 2) how these virus infections influence human gene expression and perturb the homeostasis of these tissues.
“Hidden” virome in the human body
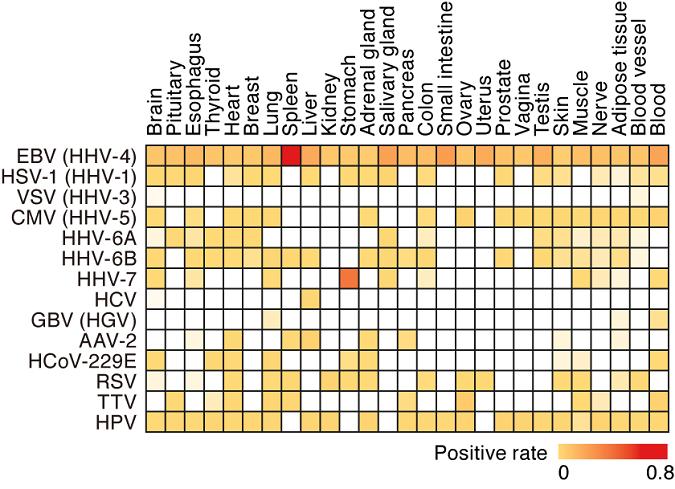
To characterize the human virome in a tissue-specific manner, the research group performed meta-transcriptomic analysis(*4) using the RNA-sequencing dataset(*5) from the Genotype-Tissue Expression (GTEx) Project(*6) with SHIROKANE, a supercomputer in IMSUT. They analyzed 8,991 RNA-sequencing datasets obtained from 51 somatic tissues from 547 individuals and successfully detected 39 viral species in at least one tissue.
The research group then investigated associations between virus infection and human gene expression and human disease onset. They detected some expected relationships; for instance, hepatitis C virus infection in the liver was strongly associated with interferon-stimulated gene upregulation and pathological findings of chronic hepatitis. Presence of herpes simplex virus type 1 in one subject’s brain was strongly associated with immune gene expression.
While torque teno virus was detected in a broad range of human tissues, it was not associated with interferon responses. Being notable in light of its association with lymphoproliferative disorders, Epstein-Barr virus infection in the spleen and blood was associated with an increase in plasma cells in healthy subjects. Human herpesvirus 7 was often detected in the stomach; intriguingly, it was associated with the proportion of human leukocytes in the stomach as well as digestive gene expression. Moreover, virus infections in local tissues was associated with systemic immune responses in circulating blood.
Associate Professor Sato emphasized the work’s importance; “To our knowledge, this study is the first comprehensive investigation of the human virome in a variety of tissues in healthy individuals through meta-transcriptomic analysis. Further investigation of the associations described here, and application of this analytical pipeline to additional datasets, will be useful to reveal the impact of viral infections on human health” .
###
Research Notes
(*1) Sequencing
Nucleotide sequencing techniques. The next generation sequencer can be used to determine nucleotide sequence information in parallel and at high speed.
(*2) Virome
The assemblage of viruses, and a part of the microbiome.
(*3) Metagenomic analyses
A method to comprehensively analyze the genomic DNA in a sample. It is mainly used to examine the microbial community in a sample.
(*4) Meta-transcriptomic analysis
A method for exhaustive analysis of the RNA present in a sample. In this study, the profiles of human gene-derived and virus-derived sequences in the RNA-seq data were comprehensively obtained.
(*5) RNA-sequencing dataset
The sequence information of the RNA present in the sample. It is mainly obtained for the purpose of quantifying gene expression (transcripts). It is also called a transcriptome.
(*6) Genotype-Tissue Expression (GTEx) project
An ongoing project in the US to build a comprehensive public resource to study tissue-specific gene expression and regulation.
About the research
Ryuichi Kumata, Jumpei Ito, Kenta Takahashi, Tadaki Suzuki, Kei Sato,2020,”A tissue level atlas of the healthy human virome”BMC Biology
DOI: 10.1186/s12915-020-00785-5
Papers
Article URL: https:/
Funding
This study was supported in part by JST CREST (to Kei Sato) including AIP challenge program (to Ryuichi Kumata); AMED J-PRIDE JP19fm0208006 (to Kei Sato); JSPS Scientific Research B JP18H02662 (to Kei Sato), Scientific Research on Innovative Areas JP16H06429 (to Kei Sato), JP16K21723 (to Kei Sato), JP17H05813 (to Kei Sato), and JP19H04826 (to Kei Sato), JSPS Research Fellow PD JP19J01713 (to Jumpei Ito).
Media Contact
Associate Professor Kei Sato
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




