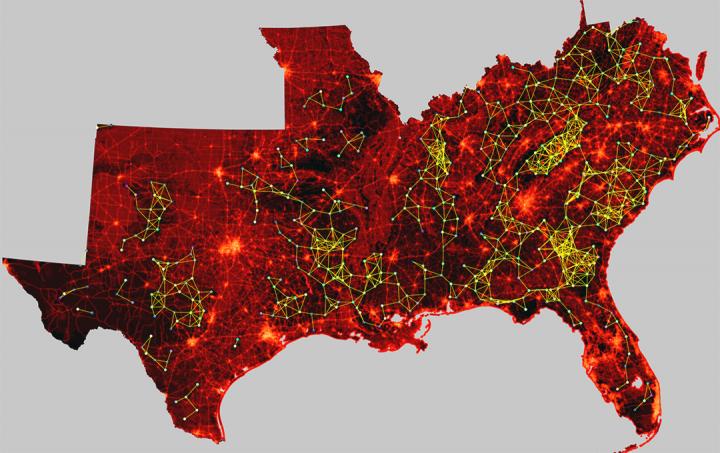
Credit: Paul Leonard / Clemson University
CLEMSON, South Carolina – A trio of Clemson University scientists has unveiled a groundbreaking computational software called "GFlow" that makes wildlife habitat connectivity modeling vastly faster, more efficient and superior in quality and scope.
After eight years of research and development, the revolutionary software was announced in the scientific journal Methods in Ecology and Evolution. Clemson University postdoctoral fellow Paul Leonard is the lead author of the article, "GFlow: software for modeling circuit theory-based connectivity at any scale." Clemson's co-authors are Rob Baldwin, the Margaret H. Lloyd-Smart State Endowed Chair in the forestry and environmental conservation department; and Edward Duffy, formerly a computational scientist in the cyberinfrastructure technology integration department who recently left the university to join BMW.
"Historically, landscape connectivity mapping has been limited in either extent or spatial resolution, largely because of the amount of time it took computers to solve the enormous equations necessary to create these models. Even using a supercomputer, it could take days, weeks or months," said Leonard, who is in the forestry and environmental conservation department along with Baldwin. "But GFlow is more than 170 times faster than any previously existing software, removing limitations in resolution and scale and providing users with a level of quality that will be far more effective in presenting the complexities of landscape networks."
Habitat connectivity maps are paired with satellite imagery to display the potential corridors used by animal populations to move between both large and small areas. Billions of bytes of data – including fine-grain satellite photographs and on-the-ground research – produce geospatial models of the movements of everything from black bears to white salamanders. These models help federal and state governments, non-governmental organizations and individual landowners redefine their conservation priorities by computationally illustrating the passageways that will need to be preserved and enhanced for animals to be able to continue to intermingle.
"The take-home from this is that you can quickly compute very complicated scenarios to show decision-makers the impacts of various outcomes," said Baldwin, whose conservation career has spanned decades throughout the United States and Canada. "You want to put a road here? Here's what happens to the map. You want to put the road over there? We'll recalculate it and show you how the map changes. GFlow is dynamic, versatile and powerful. It's a game-changer in a variety of ways."
When Leonard began his initial work under Baldwin's tutorship, the existing software used for habitat connectivity mapping was slow, inefficient and consumed enormous amounts of computer memory. Leonard soon realized he would need the expertise of a computational scientist to overcome these frustrating limitations. Thus, his collaboration with Duffy began – and both ended up spending countless hours in front of their computer screens, synthesizing Leonard's ecological know-how with Duffy's cyber skills. The end result? A large-scale map that once would have taken more than a year to generate now takes just a few days.
"Until GFlow, the software available for ecologists was poorly conceived in terms of speed and memory usage," said Duffy, who was the lead developer of the new software. "So I rewrote the code from scratch and reduced individual calculations from about 30 minutes to three seconds. And I also significantly reduced the amount of memory generated by the program. In the old code, one project we worked on took up 90 gigabytes of memory. With GFlow, only about 20 gigabytes would be needed. It's most efficient when used in conjunction with a supercomputer, but it even works in a more limited capacity on desktop computers."
GFlow will enable scientists to solve ecological problems that span large landscapes. But in addition to helping animals survive and thrive, GFlow can also be used for human health and well-being. For instance, GFlow has the capacity to monitor the spread of the Zika virus by documenting the location of each new case and then predicting its potential spread to previously uninfected areas.
"This software can monitor the flow of any natural phenomenon across space where there is heterogeneous movement that is based on some resistance to this movement," Leonard said. "Besides Zika, there are other health and disease patterns that can be modeled using GFlow. And also other natural phenomena, such as the spread of wildfire in the southeastern United States and other areas around the country. We can parameterize wind strength and shifts, how much fuel is on the ground and calculate the spread across really large areas. So we're examining all these possibilities and are open to collaboration with other domain experts who might be interested in using GFlow."
Additional contributors to Friday's journal article were Brad McRae, a senior landscape ecologist for The Nature Conservancy; and Viral Shah and Tanmay Mohapatra of Julia Computing, a privately held company. Ron Sutherland and the Wildlands Network provided valuable data that was used extensively during the development of GFlow
"Collaboration has played a huge role in this," Baldwin said. "We've worked across departments. We've worked across boundaries. And without Clemson's investment in the Palmetto Cluster supercomputer, none of this would have been possible. This collaboration has improved spatial modeling for ecological processes in time and space. And because it's so computationally efficient, it can be done for extremely large areas – regions, nations, continents or possibly even the entire planet – in unprecedented detail."
###
Media Contact
Jim Melvin
[email protected]
864-656-2268
@researchcu
http://www.clemson.edu
############
Story Source: Materials provided by Scienmag